On this put up we’ll establish and visualise completely different clusters of most cancers sorts by analysing illness ontology as a data graph. Particularly we’ll arrange neo4j in a docker container, import the ontology, generate graph clusters and embeddings, earlier than utilizing dimension discount to plot these clusters and derive some insights. Though we’re utilizing `disease_ontology` for example, the identical steps can be utilized to discover any ontology or graph database.
In a graph database, fairly than storing knowledge as rows (like a spreadsheet or relational database) knowledge is saved as nodes and relationships between nodes. For instance within the determine beneath we see that melanoma and carcinoma are SubCategories Of cell kind most cancers tumour (proven by the SCO relationship). With this type of knowledge we are able to clearly see that melanoma and carcinoma are associated though this isn’t explicitly said within the knowledge.
Ontologies are a formalised set of ideas and relationships between these ideas. They’re much simpler for computer systems to parse than free textual content and due to this fact simpler to extract which means from. Ontologies are extensively utilized in organic sciences and chances are you’ll discover an ontology you’re concerned about at https://obofoundry.org/. Right here we’re specializing in the illness ontology which exhibits how various kinds of ailments relate to one another.
Neo4j is a software for managing, querying and analysing graph databases. To make it simpler to arrange we’ll use a docker container.
docker run
-it - rm
- publish=7474:7474 - publish=7687:7687
- env NEO4J_AUTH=neo4j/123456789
- env NEO4J_PLUGINS='["graph-data-science","apoc","n10s"]'
neo4j:5.17.0
Within the above command the `-publish` flags set ports to let python question the database straight and allow us to entry it by a browser. The `NEO4J_PLUGINS` argument specifies which plugins to put in. Sadly, the home windows docker picture doesn’t appear to have the ability to deal with the set up, so to comply with alongside you’ll want to put in neo4j desktop manually. Don’t fear although, the opposite steps ought to all nonetheless give you the results you want.
Whereas neo4j is working you may entry your database by going to http://localhost:7474/ in your browser, or you should utilize the python driver to attach as beneath. Word that we’re utilizing the port we printed with our docker command above and we’re authenticating with the username and password we additionally outlined above.
URI = "bolt://localhost:7687"
AUTH = ("neo4j", "123456789")
driver = GraphDatabase.driver(URI, auth=AUTH)
driver.verify_connectivity()
After you have your neo4j database arrange, it’s time to get some knowledge. The neo4j plug-in n10s is constructed to import and deal with ontologies; you should utilize it to embed your knowledge into an current ontology or to discover the ontology itself. With the cypher instructions beneath we first set some configs to make the outcomes cleaner, then we arrange a uniqueness constraint, lastly we really import illness ontology.
CALL n10s.graphconfig.init({ handleVocabUris: "IGNORE" });
CREATE CONSTRAINT n10s_unique_uri FOR (r:Useful resource) REQUIRE r.uri IS UNIQUE;
CALL n10s.onto.import.fetch(http://purl.obolibrary.org/obo/doid.owl, RDF/XML);
To see how this may be performed with the python driver, take a look at the complete code right here https://github.com/DAWells/do_onto/blob/major/import_ontology.py
Now that we’ve imported the ontology you may discover it by opening http://localhost:7474/ in your internet browser. This allows you to discover somewhat of your ontology manually, however we’re within the greater image so lets do some evaluation. Particularly we’ll do Louvain clustering and generate quick random projection embeddings.
Louvain clustering is a clustering algorithm for networks like this. Briefly, it identifies units of nodes which might be extra linked to one another than they’re to the broader set of nodes; this set is then outlined as a cluster. When utilized to an ontology it’s a quick solution to establish a set of associated ideas. Quick random projection however produces an embedding for every node, i.e. a numeric vector the place extra related nodes have extra related vectors. With these instruments we are able to establish which ailments are related and quantify that similarity.
To generate embeddings and clusters we’ve to “undertaking” the components of our graph that we’re concerned about. As a result of ontologies are usually very giant, this subsetting is a straightforward solution to velocity up computation and keep away from reminiscence errors. On this instance we’re solely concerned about cancers and never every other kind of illness. We do that with the cypher question beneath; we match the node with the label “most cancers” and any node that’s associated to this by a number of SCO or SCO_RESTRICTION relationships. As a result of we need to embrace the relationships between most cancers sorts we’ve a second MATCH question that returns the linked most cancers nodes and their relationships.
MATCH (most cancers:Class {label:"most cancers"})<-[:SCO|SCO_RESTRICTION *1..]-(n:Class)
WITH n
MATCH (n)-[:SCO|SCO_RESTRICTION]->(m:Class)
WITH gds.graph.undertaking(
"proj", n, m, {}, {undirectedRelationshipTypes: ['*']}
) AS g
RETURN g.graphName AS graph, g.nodeCount AS nodes, g.relationshipCount AS rels
As soon as we’ve the projection (which we’ve referred to as “proj”) we are able to calculate the clusters and embeddings and write them again to the unique graph. Lastly by querying the graph we are able to get the brand new embeddings and clusters for every most cancers kind which we are able to export to a csv file.
CALL gds.fastRP.write(
'proj',
{embeddingDimension: 128, randomSeed: 42, writeProperty: 'embedding'}
) YIELD nodePropertiesWrittenCALL gds.louvain.write(
"proj",
{writeProperty: "louvain"}
) YIELD communityCount
MATCH (most cancers:Class {label:"most cancers"})<-[:SCO|SCO_RESTRICTION *0..]-(n)
RETURN DISTINCT
n.label as label,
n.embedding as embedding,
n.louvain as louvain
Let’s take a look at a few of these clusters to see which sort of cancers are grouped collectively. After we’ve loaded the exported knowledge right into a pandas dataframe in python we are able to examine particular person clusters.
Cluster 2168 is a set of pancreatic cancers.
nodes[nodes.louvain == 2168]["label"].tolist()
#array(['"islet cell tumor"',
# '"non-functioning pancreatic endocrine tumor"',
# '"pancreatic ACTH hormone producing tumor"',
# '"pancreatic somatostatinoma"',
# '"pancreatic vasoactive intestinal peptide producing tumor"',
# '"pancreatic gastrinoma"', '"pancreatic delta cell neoplasm"',
# '"pancreatic endocrine carcinoma"',
# '"pancreatic non-functioning delta cell tumor"'], dtype=object)
Cluster 174 is a bigger group of cancers however principally carcinomas.
nodes[nodes.louvain == 174]["label"]
#array(['"head and neck cancer"', '"glottis carcinoma"',
# '"head and neck carcinoma"', '"squamous cell carcinoma"',
#...
# '"pancreatic squamous cell carcinoma"',
# '"pancreatic adenosquamous carcinoma"',
#...
# '"mixed epithelial/mesenchymal metaplastic breast carcinoma"',
# '"breast mucoepidermoid carcinoma"'], dtype=object)p
These are smart groupings, based mostly on both organ or most cancers kind, and can be helpful for visualization. The embeddings however are nonetheless too excessive dimensional to be visualised meaningfully. Happily, TSNE is a really helpful technique for dimension discount. Right here, we use TSNE to scale back the embedding from 128 dimensions right down to 2, whereas nonetheless protecting intently associated nodes shut collectively. We will confirm that this has labored by plotting these two dimensions as a scatter plot and colouring by the Louvain clusters. If these two strategies agree we must always see nodes clustering by color.
from sklearn.manifold import TSNEnodes = pd.read_csv("export.csv")
nodes['louvain'] = pd.Categorical(nodes.louvain)
embedding = nodes.embedding.apply(lambda x: ast.literal_eval(x))
embedding = embedding.tolist()
embedding = pd.DataFrame(embedding)
tsne = TSNE()
X = tsne.fit_transform(embedding)
fig, axes = plt.subplots()
axes.scatter(
X[:,0],
X[:,1],
c = cm.tab20(Normalize()(nodes['louvain'].cat.codes))
)
plt.present()
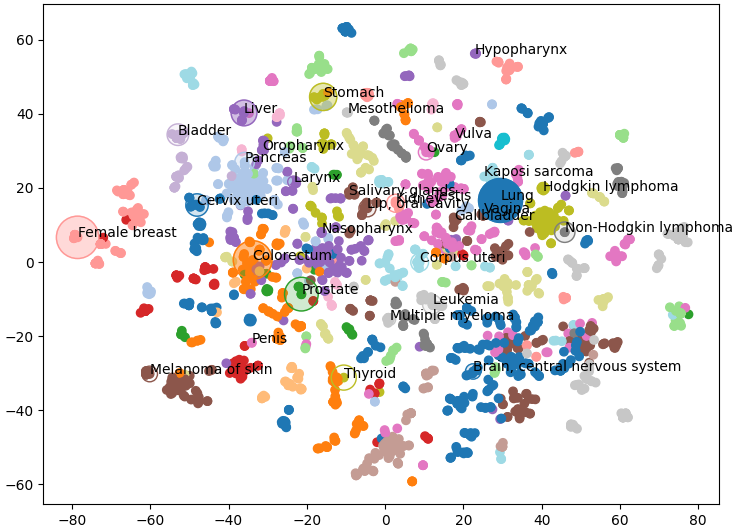
Which is strictly what we see, related sorts of most cancers are grouped collectively and visual as clusters of a single color. Word that some nodes of a single color are very far aside, it is because we’re having to reuse some colors as there are 29 clusters and solely 20 colors. This offers us an important overview of the construction of our data graph, however we are able to additionally add our personal knowledge.
Under we plot the frequency of most cancers kind as node measurement and the mortality price because the opacity (Bray et al 2024). I solely had entry to this knowledge for a couple of of the most cancers sorts so I’ve solely plotted these nodes. Under we are able to see that liver most cancers doesn’t have an particularly excessive incidence over all. Nonetheless, incidence charges of liver most cancers are a lot larger than different cancers inside its cluster (proven in purple) like oropharynx, larynx, and nasopharynx.
Right here we’ve used the illness ontology to group completely different cancers into clusters which provides us the context to check these ailments. Hopefully this little undertaking has proven you methods to visually discover an ontology and add that data to your personal knowledge.
You’ll be able to take a look at the complete code for this undertaking at https://github.com/DAWells/do_onto.
Bray, F., Laversanne, M., Sung, H., Ferlay, J., Siegel, R. L., Soerjomataram, I., & Jemal, A. (2024). International most cancers statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 nations. CA: a most cancers journal for clinicians, 74(3), 229–263.