Digital pathology is crucial for the prognosis and remedy of most cancers, enjoying a crucial function in healthcare supply and pharmaceutical analysis and improvement. Pathology historically depends closely on pathologist experience and expertise to conduct meticulous examination of tissue samples to establish abnormalities. Nevertheless, the growing complexity and quantity of instances necessitate superior instruments to help pathologists in making quicker, extra correct diagnoses.
The digitization of pathology slides, often known as entire slide pictures (WSIs), gave rise to the brand new subject of computational pathology. By making use of AI to those digitized WSIs, researchers are working to unlock new insights and improve present annotations workflows. A pivotal development within the subject of computational pathology has been the emergence of large-scale deep neural community architectures, often known as basis fashions (FMs). These fashions are educated utilizing self-supervised studying algorithms on expansive datasets, enabling them to seize a complete repertoire of visible representations and patterns inherent inside pathology pictures. The facility of FMs lies of their means to be taught strong and generalizable information embeddings that may be successfully transferred and fine-tuned for all kinds of downstream duties, starting from automated illness detection and tissue characterization to quantitative biomarker evaluation and pathological subtyping.
Not too long ago, French startup Bioptimus introduced the launch of a brand new pathology imaginative and prescient FM: H-optimus-0, the world’s largest publicly out there FM for pathology. With 1.1 billion parameters, H-optimus-0 was educated on a proprietary dataset of a number of tons of of thousands and thousands of pictures extracted from over 500,000 histopathology slides. This units a brand new benchmark for state-of-the-art efficiency in crucial medical diagnostic duties, from figuring out cancerous cells to detecting genetic abnormalities in tumors.
The current addition of H-optimus-0 to Amazon SageMaker JumpStart marks a big milestone in making superior AI capabilities accessible to healthcare organizations. This highly effective FM, with its complete coaching on over 500,000 histopathology slides, represents a priceless instrument for organizations seeking to improve their digital pathology workflows.
On this submit, we exhibit find out how to use H-optimus-0 for 2 widespread digital pathology duties: patch-level evaluation for detailed tissue examination, and slide-level evaluation for broader diagnostic evaluation. By means of sensible examples, we present you find out how to adapt this FM to those particular use instances whereas optimizing computational assets.
Resolution overview
Our resolution makes use of the AWS built-in ecosystem to create an environment friendly scalable pipeline for digital pathology AI workflows. The structure combines the next providers:
The next diagram illustrates the answer structure for coaching and deploying fine-tuned FMs utilizing H-optimus-0.

This diagram illustrates the answer structure for coaching and deploying fine-tuned FMs utilizing H-optimus-0
This submit supplies instance scripts and coaching notebooks within the following GitHub repository.
Conditions
We assume you could have entry to and are authenticated in an AWS account. The AWS CloudFormation template for this resolution makes use of t3.medium situations to host the SageMaker pocket book. Characteristic extraction makes use of g5.2xlarge occasion varieties powered by NVIDIA T4 GPU examined within the us-west-2 AWS Area. Coaching jobs are run on p3.2xlarge and g5.2xlarge situations. Examine your AWS service quotas to be sure you have ample entry to those occasion varieties.
Create the AWS infrastructure
To get began with pathology AI workflows, we use AWS CloudFormation to automate the setup of our core infrastructure. The offered infra-stack.yml template creates a whole setting prepared for mannequin fine-tuning and coaching.
Our CloudFormation stack configures a safe networking setting utilizing Amazon Digital Non-public Cloud (Amazon VPC), establishing each private and non-private subnets with acceptable gateways for web connectivity. Inside this community, it creates an EFS file system to effectively retailer and serve massive pathology slide pictures. The stack additionally provisions a SageMaker pocket book occasion that mechanically connects to the EFS storage, offering seamless entry to coaching information.
The template handles all crucial safety configurations, together with AWS Identification and Entry Administration (IAM) roles. When deploying the stack, make observe of the personal subnet and safety group identifiers; you’ll need to verify your coaching jobs can entry the EFS information storage.
For detailed setup directions and configuration choices, discuss with the README in our GitHub repository.
Use FMs for patch-level prediction duties
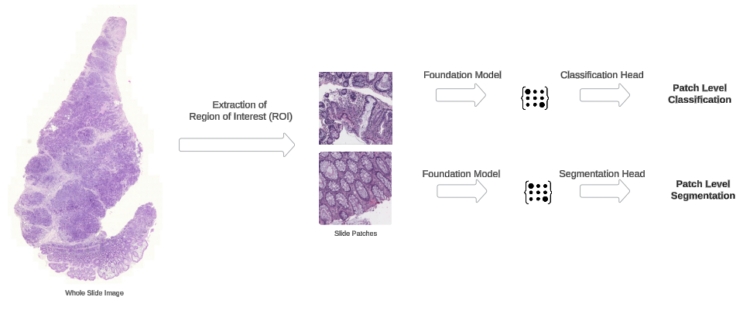
Patch-level evaluation is key to digital pathology AI workflows. As an alternative of processing whole WSIs that may exceed a number of gigabytes, patch-level evaluation focuses on particular tissue areas. This focused strategy allows environment friendly useful resource utilization and quicker mannequin improvement cycles. The next diagram illustrates the workflow of patch-level prediction duties on a WSI.

This diagram illustrates the workflow of patch-level prediction duties on a WSI
Classification activity: MHIST dataset
We exhibit patch-level classification utilizing the MHIST dataset, which comprises colorectal polyp pictures. Early detection of probably cancerous polyps straight impacts affected person survival charges, making this a clinically related use case. By including a easy classification head on high of H-optimus-0’s pretrained options and utilizing linear probing, we obtain 83% accuracy. The implementation makes use of Amazon EFS for environment friendly information streaming and p3.2xlarge situations for optimum GPU utilization.
To entry the MHIST dataset, submit an information request via their portal to acquire the annotations.csv file and pictures.zip file. Our repository features a download_mhist.sh script that mechanically downloads and organizes the info in your EFS storage.
Segmentation activity: Lizard dataset
For our second patch-level activity, we exhibit nuclear segmentation utilizing the Lizard dataset, which requires exact pixel-level predictions of nuclear boundaries in colon tissue. We adapt H-optimus-0 for segmentation by including a Mask2Former ViT adapter head, permitting the mannequin to generate detailed segmentation masks whereas utilizing the FM’s highly effective function extraction capabilities.
The Lizard dataset is out there on Kaggle, and our repository consists of scripts to mechanically obtain and put together the info for coaching. The segmentation implementation runs on g5.16xlarge situations to deal with the computational calls for of pixel-level predictions.
Use FMs for WSI-level duties
Analyzing whole WSIs presents distinctive challenges attributable to their huge measurement, typically exceeding 50,000 x 50,000 pixels. To deal with this, we implement a number of occasion studying (MIL), which treats every WSI as a group of smaller patches. Our attention-based MIL strategy mechanically learns which areas are most related for the ultimate prediction. The next diagram illustrates the workflow for WSI-level prediction duties utilizing MIL.

This diagram illustrates the workflow for WSI-level prediction duties utilizing MIL
WSI processing pipeline
Our implementation optimizes WSI evaluation via the next strategies:
- Clever patching – We use the GPU-accelerated CuCIM library to effectively load WSIs and apply Canny edge detection to establish and extract solely tissue-containing areas
- Characteristic extraction – The chosen patches are processed in parallel utilizing GPU acceleration, with options saved in space-efficient HDF5 format for downstream evaluation
MSI standing prediction
We exhibit our WSI pipeline by predicting microsatellite instability (MSI) standing, an important biomarker that guides immunotherapy choices in most cancers remedy. The TCGA-COAD dataset used for this activity will be accessed via the GDC Knowledge Portal, and our repository supplies detailed directions for downloading the WSIs and corresponding MSI labels.
Clear up
After you’ve completed, don’t overlook to delete the related assets (Amazon EFS storage and SageMaker pocket book situations) to keep away from sudden prices.
Conclusion
On this submit, we demonstrated how you need to use AWS providers to construct scalable digital pathology AI workflows utilizing the H-optimus-0 FM. By means of sensible examples of each patch-level duties (MHIST classification and Lizard nuclear segmentation) and WSI evaluation (MSI standing prediction), we confirmed find out how to effectively deal with the distinctive challenges of computational pathology.
Our implementation highlights the seamless integration between AWS providers for dealing with large-scale pathology information processing. Though we used Amazon EFS for this demonstration to allow high-throughput coaching workflows, manufacturing deployments would possibly think about AWS HealthImaging for long-term storage of medical imaging information.
We hope this pipeline serves as a place to begin on your personal pathology AI initiatives. The offered GitHub repository comprises the mandatory elements that will help you start constructing and scaling pathology workflows on your particular use instances. You may clone the repository and arrange the infrastructure utilizing the offered CloudFormation template. Then strive fine-tuning H-optimus-0 by yourself pathology datasets and downstream duties and evaluate the outcomes together with your present strategies.
We’d love to listen to about your experiences and insights. Attain out to us or contribute to the publicly out there FMs to assist advance the sphere of computational pathology.
In regards to the Authors
 Pierre de Malliard is a Senior AI/ML Options Architect at Amazon Internet Providers and helps clients within the healthcare and life sciences business. In his free time, Pierre enjoys snowboarding and exploring the New York meals scene.
Pierre de Malliard is a Senior AI/ML Options Architect at Amazon Internet Providers and helps clients within the healthcare and life sciences business. In his free time, Pierre enjoys snowboarding and exploring the New York meals scene.
 Christopher is a senior companion account supervisor at Amazon Internet Providers (AWS), serving to unbiased software program distributors (ISVs) innovate, construct, and co-sell cloud-based healthcare software-as-a-service (SaaS) options in public sector. A part of the Healthcare and Life Sciences Technical Discipline Group (TFC), Christopher goals to speed up the digitization and utilization of healthcare information to drive improved outcomes and customized care supply.
Christopher is a senior companion account supervisor at Amazon Internet Providers (AWS), serving to unbiased software program distributors (ISVs) innovate, construct, and co-sell cloud-based healthcare software-as-a-service (SaaS) options in public sector. A part of the Healthcare and Life Sciences Technical Discipline Group (TFC), Christopher goals to speed up the digitization and utilization of healthcare information to drive improved outcomes and customized care supply.