This submit was written with Audra Devoto, Owen Janson, and Christopher Brown of Metagenomi, and Adam Perry of Tennex.
A promising technique to enhance the in depth pure variety of excessive worth enzymes is to make use of generative AI, particularly protein language fashions (pLMs), skilled on identified enzymes to create orders of magnitude extra predicted examples of given enzyme courses. Increasing pure enzyme variety by generative AI has many benefits, together with offering entry to quite a few enzyme variants that may provide enhanced stability, specificity, or efficacy in human cells—however excessive throughput era might be pricey relying on the dimensions of the mannequin used and the variety of enzyme variants wanted.
At Metagenomi, we’re creating probably healing therapeutics with proprietary CRISPR gene enhancing enzymes. We use the in depth pure variety of enzymes in our database (MGXdb) to determine pure enzyme candidates, and to coach protein language fashions used for generative AI. Growth of pure enzyme courses with generative AI permits us to entry extra variants of a given enzyme class, that are filtered with multi-model workflows to foretell key enzyme traits and used to allow protein engineering campaigns to enhance enzyme efficiency in a given context.
On this weblog submit, we element strategies for value discount of excessive throughput protein generative AI workflows by implementing the Progen2 mannequin on AWS Inferentia, which enabled excessive throughput era of enzyme variants with as much as 56% decrease value on AWS Batch and Amazon Elastic Compute Cloud (Amazon EC2) Spot Situations. This work was performed in partnership with the AWS Neuron crew and engineers at Tennex.
Progen2 on AWS Inferentia
PyTorch fashions can use AWS neuron cores as accelerators, which led us to make use of AWS Inferentia powered EC2 Inf2 occasion sorts for our high-throughput protein design workflow to make use of their cost-effectiveness and better availability as Spot Situations. We selected the autoregressive transformer mannequin Progen2 to implement on EC2 Inf2 occasion sorts, as a result of it met our wants for top throughput artificial protein era from advantageous tuned fashions primarily based on earlier work working Progen2 on EC2 situations with NVIDIA L40S GPUs (g6e.xlarge), and since there’s established assist in Neuron for transformer decoder sort fashions. To implement Progen2 on EC2 Inf2 situations, we traced customized Progen2 checkpoints skilled on proprietary enzymes utilizing the bucketing method. Tracing out fashions to a number of sizes optimizes for mannequin efficiency by producing sequences on consecutively bigger traced fashions, passing the output of the earlier mannequin as a immediate to the following one, which minimizes the inference time required to generate every token.
Nevertheless, the tracing and bucketing method used to allow Progen2 to run on EC2 Inf2 occasion introduces some adjustments that might influence mannequin accuracy. For instance, Progen2-base was not skilled with padding tokens, which should be added to enter tensors when working on EC2 Inf2 situations. To check the results of our tracing and bucketing method, we in contrast the perplexity and sequence lengths of sequences generated utilizing the progen2-base mannequin on EC2 Inf2 situations to these utilizing the native implementation of the progen2-base mannequin on NVIDIA GPUs.
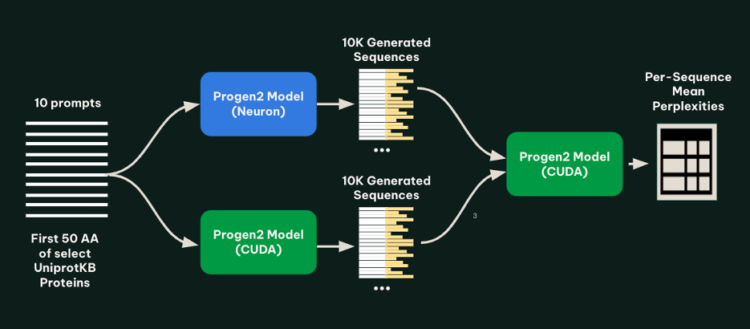
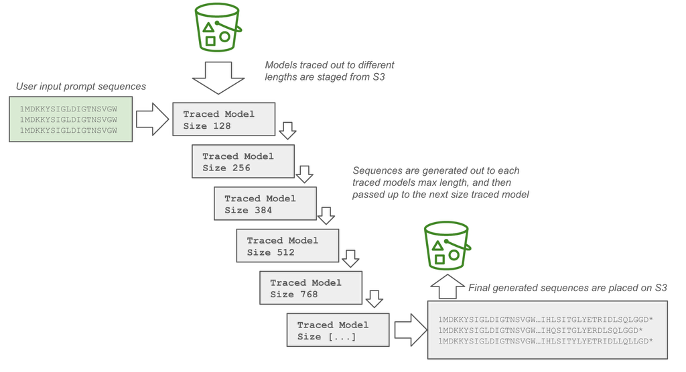
To check the fashions, we generated 1,000 protein sequences for every of 10 prompts below each the tracing and bucketing (AWS AI Chip Inferentia) implementation and the native (NVIDIA GPU) implementation. The set of prompts was created by drawing 10 well-characterized proteins from UniprotKB, and truncating every to the primary 50 amino acids. To verify for abnormalities within the generated sequences, all generated sequences from each fashions have been then run by a ahead cross of the native implementation of progen2-base, and the imply perplexity of every sequence was calculated. The next determine illustrates this methodology.
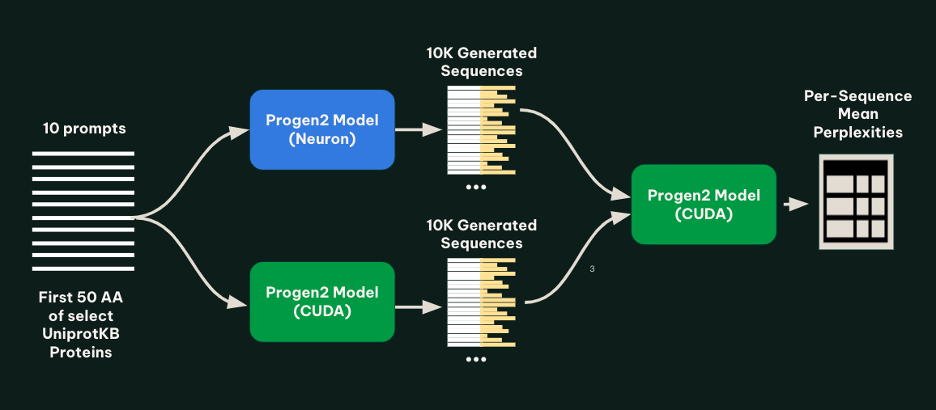
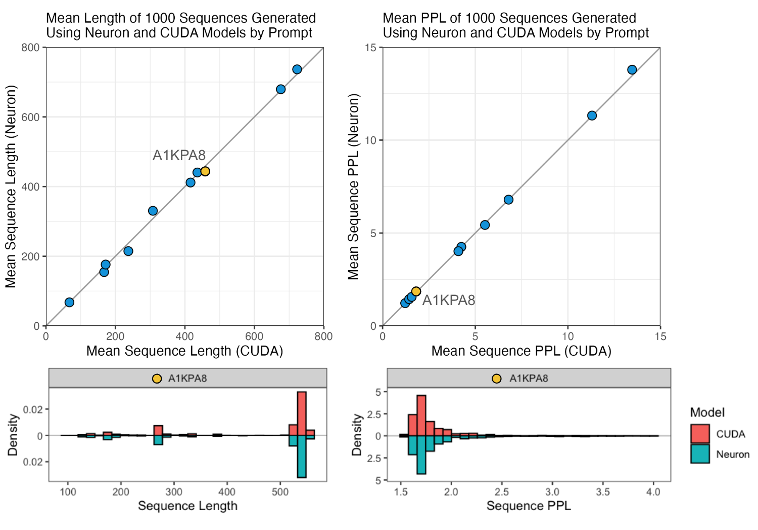
As proven within the following determine, we discovered that for every immediate, the lengths and perplexities of sequences generated utilizing the tracing and bucketing implementation and native implementation seemed related.
Scaled inference on AWS Batch
With the essential inference logic utilizing Progen2 on EC2 Inf2 situations labored out, the following step was to massively scale inference throughout a big fleet of compute nodes. AWS Batch was the perfect service to scale this workflow, as a result of it could possibly effectively run a whole lot of 1000’s of batch computing jobs and dynamically provision to the optimum amount and sort of compute assets (akin to Amazon EC2 On-Demand or Spot Situations) primarily based on the amount and useful resource necessities of the submitted jobs.
Progen2 was applied as a batch workload following finest practices. Jobs are submitted by a person, and run on a devoted compute atmosphere that orchestrates Amazon EC2 inf2.xlarge Spot Situations. Customized docker containers are saved on the Amazon Amazon Elastic Container Registry (Amazon ECR). Fashions are pulled down from Amazon Easy Storage Service (Amazon S3) by every job, and generated sequences within the type of a FASTA file are positioned on Amazon S3 on the finish of every job. Optionally, Nextflow can be utilized to orchestrate jobs, deal with computerized spot retries, and automate downstream or upstream duties. The next determine illustrates the answer structure.
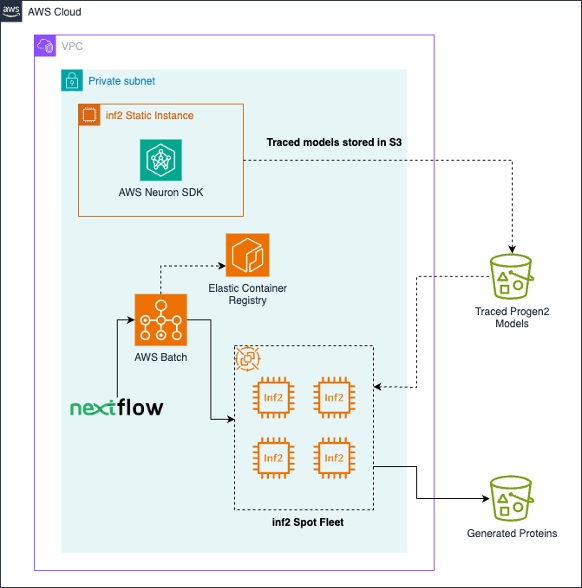
As proven within the following determine, inside every particular person batch job, sequences are generated by first loading the smallest bucket measurement from accessible traced fashions. Sequences are generated out to the max tokens for that bucket, and sequences that have been generated with a cease codon or a begin codon are dropped. The remaining unfinished sequences are handed as much as subsequently bigger bucket sizes for additional era, till the parameter max_length is reached. Tensors are stacked and decoded on the finish and written to a FASTA file that’s positioned on Amazon S3.
The next is an instance configuration sketch for organising an AWS Batch atmosphere utilizing EC2 Inf2 situations that may run Progen2-neuron workloads.
DISCLAIMER: That is pattern configuration for non-production utilization. It is best to work together with your safety and authorized groups to stick to your organizational safety, regulatory, and compliance necessities earlier than deployment.
Stipulations
Earlier than implementing this configuration, be sure you have the next stipulations in place:
The following is an example Dockerfile for containerizing Progen2-neuron. This container image builds on Amazon Linux 2023 and includes the necessary components for running Progen2 on AWS Neuron—including the Neuron SDK, Python dependencies, PyTorch, and Transformers libraries. It also configures offline mode for Hugging Face operations and includes the required sequence generation scripts.
The next is an instance generate_sequences.sh that orchestrates the sequence era workflow for Progen2 on EC2 Inf2 situations.
Price comparisons
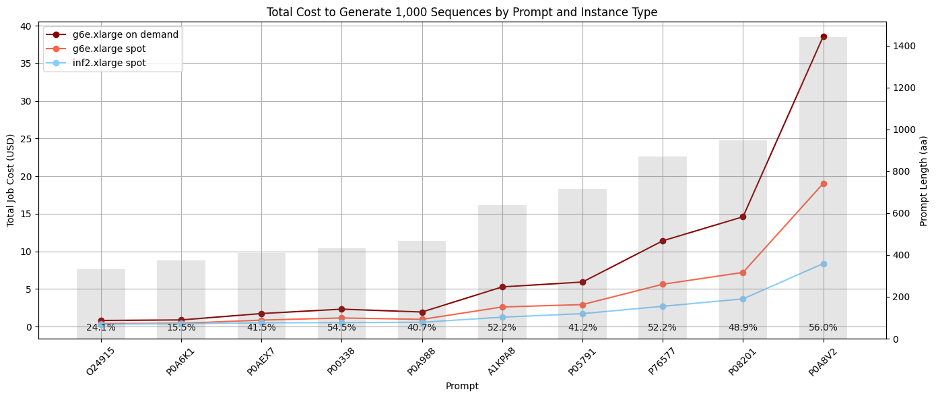
The first objective of this venture was to decrease the price of producing protein sequences with Progen2, in order that we may use this mannequin to considerably develop the variety of a number of enzyme courses. To match the price of producing sequences utilizing each companies, we generated 10,000 sequences primarily based on prompts derived from 10 widespread sequences in UniProtKB, utilizing a temperature of 1.0. Batch jobs have been run in parallel, with every job producing 100 sequences for a single immediate. We noticed that the implementation of Progen2 on EC2 Inf2 Spot Situations was considerably cheaper than implementation on Amazon EC2 G6e Spot Situations for longer sequences, representing financial savings of as much as 56%. These value estimates embrace anticipated Amazon EC2 Spot interruption frequencies of 20% for Amazon EC2 g6e.xlarge situations powered by NVIDIA L40S Tensor Core GPUs and 5% for EC2 inf2.xlarge situations. The next determine illustrates complete value the place grey bars signify the typical size of generated sequences.
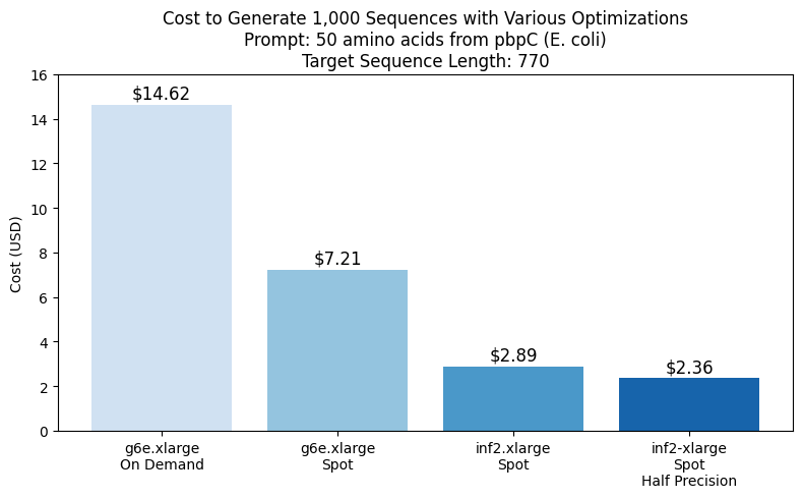
Further value financial savings might be achieved by working jobs at half precision, which appeared to supply equal outcomes, as proven within the following determine.
You will need to be aware that as a result of the time it takes so as to add new tokens to the sequence throughout era scales quadratically, the time and value to generate sequences is extremely depending on the forms of sequences you’re attempting to generate. General, we noticed that the price to generate sequences is determined by the precise mannequin checkpoint used and the distribution of sequence lengths the mannequin generated. For value estimates, we suggest producing a small subset of enzymes together with your chosen mannequin and extrapolating prices for a bigger era set, as a substitute of attempting to calculate primarily based on prices for earlier experiments.
Scaling era to tens of millions of proteins
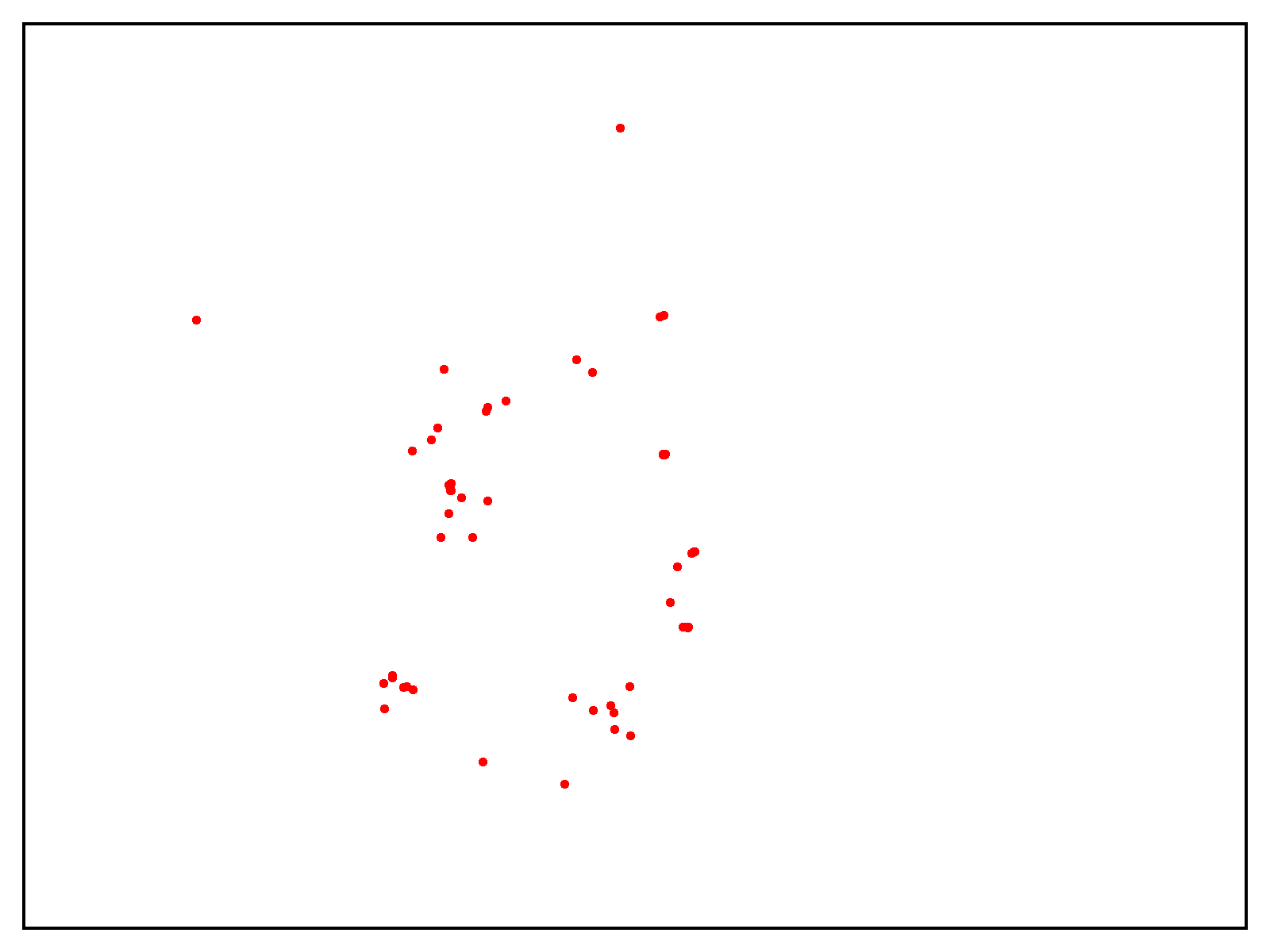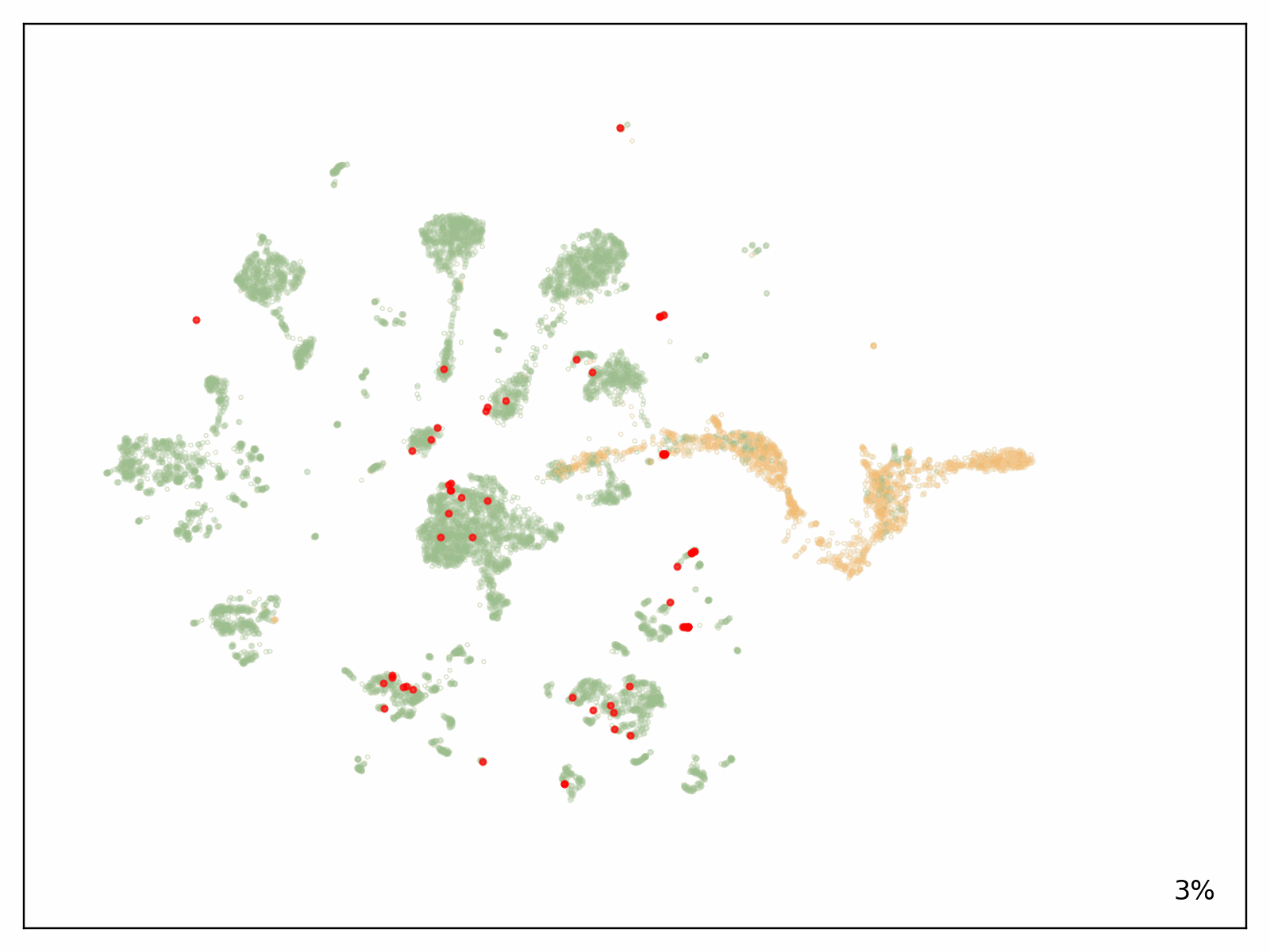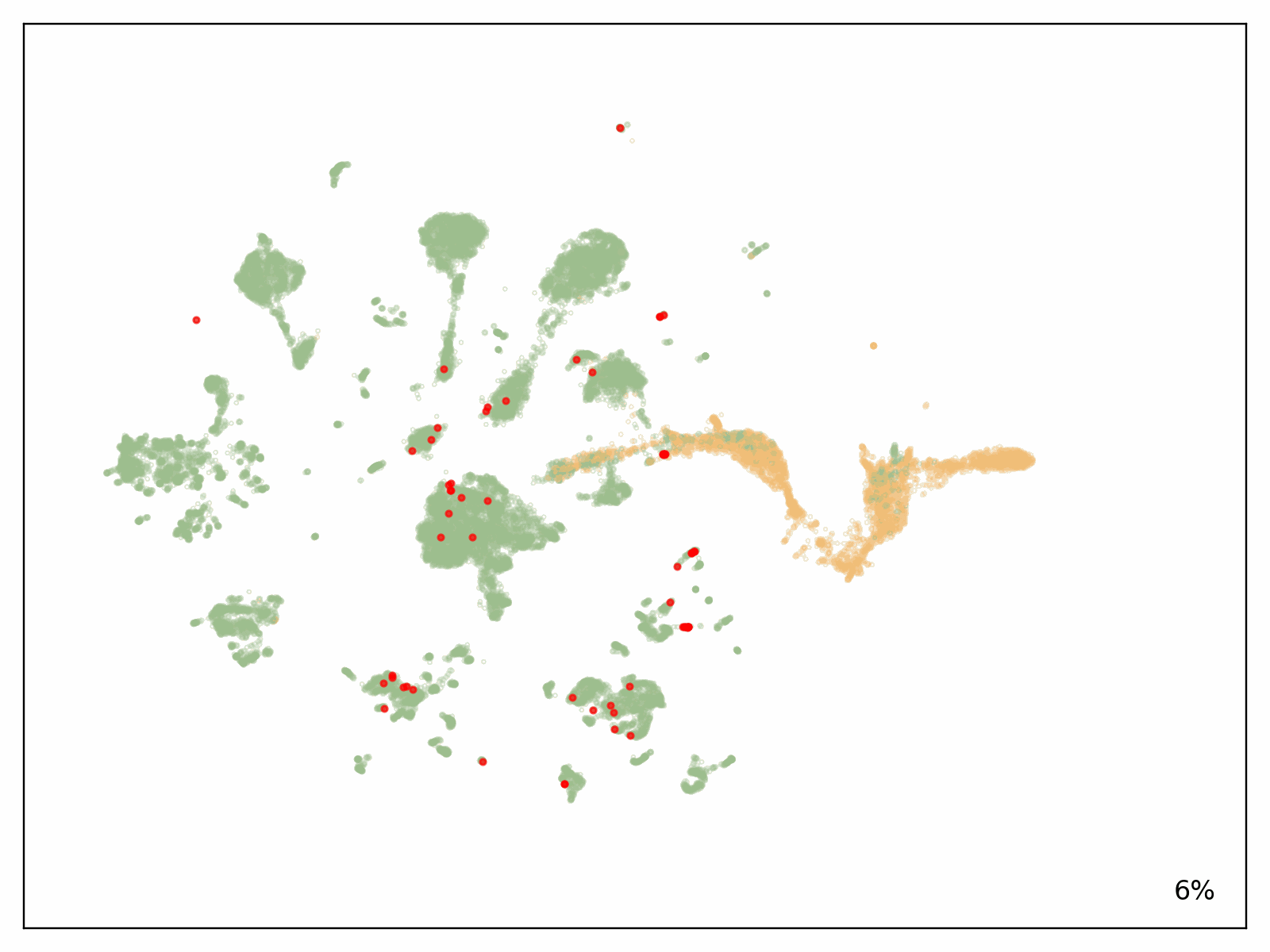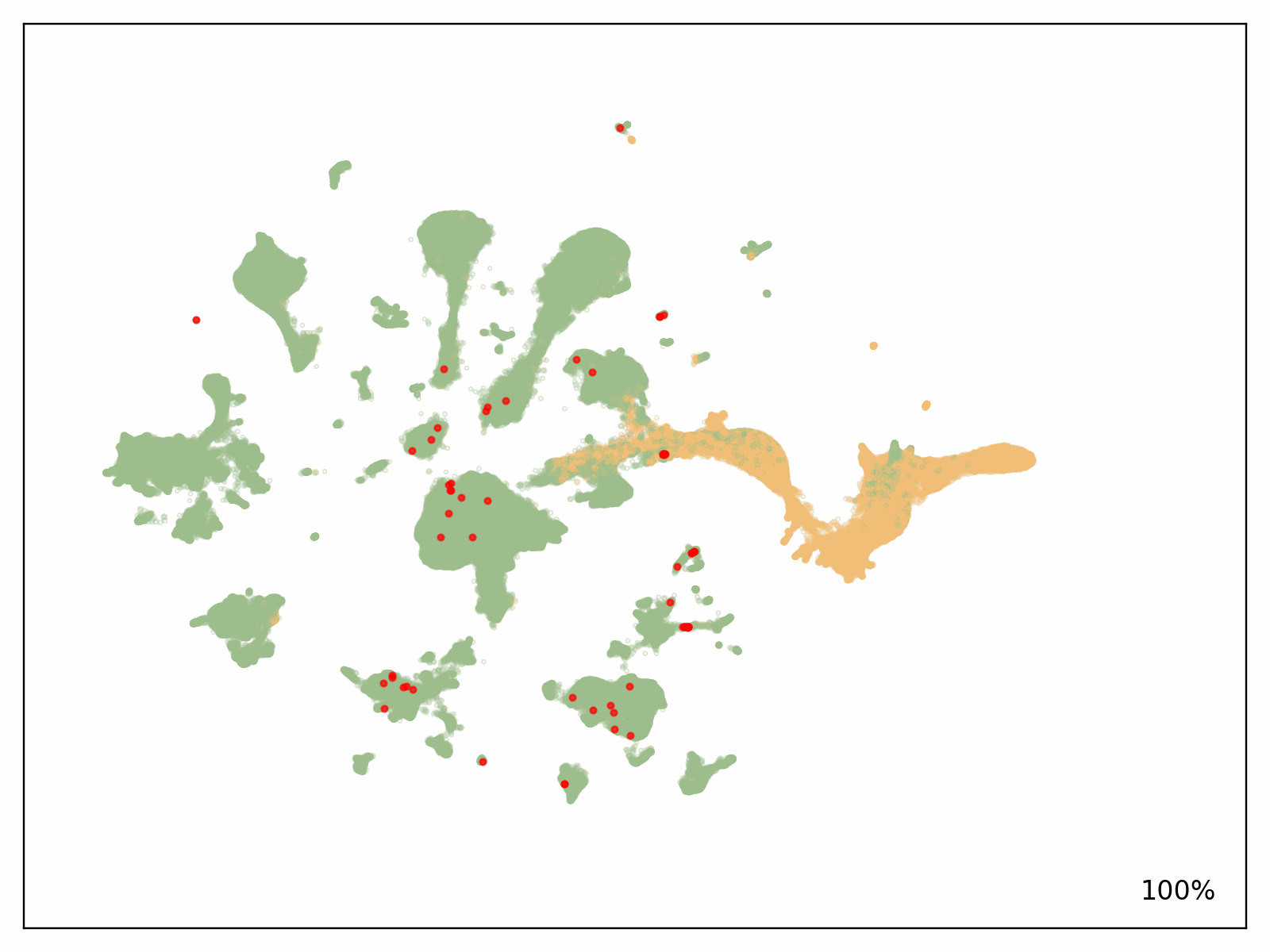
To check the scalability of our answer, Metagenomi advantageous tuned a mannequin on pure examples of a priceless however uncommon class of enzymes sourced from Metagenomi’s massive, proprietary database of metagenomics knowledge. The mannequin was advantageous tuned utilizing conventional GPU situations, then traced onto AWS AI chips Inferentia. Utilizing our batch and Nextflow pipeline, we launched batch jobs to generate effectively over 1 million enzymes, various era parameters between jobs to check the impact of various sampling strategies, temperatures, and precisions. The whole compute value of era utilizing our optimized AWS AI pipeline, together with prices incurred from EC2 Inf2 Spot retries, was $2,613 (see previous be aware on estimating prices on your workloads). Generated sequences have been validated with a pipeline that used a mixture of AI and conventional sequence validation strategies. Sequences have been dereplicated utilizing mmseqs, filtered for acceptable size, checked for correct area buildings with hmmsearch, folded utilizing ESMFold, and embedded utilizing AMPLIFY_350M. Constructions have been used for comparability to identified enzymes within the class, and embeddings have been used to validate intrinsic enzyme health. Outcomes of the era are proven within the following determine, with a number of hundred thousand generative AI enzymes plotted within the embedding area. Pure, characterised enzymes used as prompts proven in pink, generative AI enzymes passing all filters are proven in inexperienced, and generative AI enzymes not passing filters proven in orange.
Conclusion
On this submit, we outlined strategies to cut back the price of large-scale protein design tasks by as much as 56% utilizing Amazon EC2 Inf situations, which has allowed Metagenomi to generate tens of millions of novel enzymes throughout a number of excessive worth protein courses utilizing fashions skilled on our proprietary protein dataset. This implementation showcases how AWS Inferentia could make large-scale protein era extra accessible and economical for biotechnology functions. To be taught extra about EC2 Inf situations and to begin implementing your personal workflows on AWS Neuron, see the AWS Neuron documentation. To learn extra about among the novel enzymes Metagenomi has found, see Metagenomi’s publications and posters.
Concerning the authors
 Audra Devoto is a Information Scientist with a background in metagenomics and a few years of expertise working with massive genomics datasets on AWS. At Metagenomi, she builds out infrastructure to assist massive scale evaluation tasks and permits discovery of novel enzymes from MGXdb.
Audra Devoto is a Information Scientist with a background in metagenomics and a few years of expertise working with massive genomics datasets on AWS. At Metagenomi, she builds out infrastructure to assist massive scale evaluation tasks and permits discovery of novel enzymes from MGXdb.
 Owen Janson is a bioinformatics Engineer at Metagenomi who focuses on constructing instruments and cloud infrastructure to assist evaluation of huge genomic datasets.
Owen Janson is a bioinformatics Engineer at Metagenomi who focuses on constructing instruments and cloud infrastructure to assist evaluation of huge genomic datasets.
 Adam Perry is a seasoned cloud architect with deep experience in AWS, the place he’s designed and automatic complicated cloud options for a whole lot of companies. As a co-founder of Tennex, he has led know-how technique, constructed customized instruments, and collaborated carefully together with his crew to assist early stage biotech corporations scale securely and effectively on cloud.
Adam Perry is a seasoned cloud architect with deep experience in AWS, the place he’s designed and automatic complicated cloud options for a whole lot of companies. As a co-founder of Tennex, he has led know-how technique, constructed customized instruments, and collaborated carefully together with his crew to assist early stage biotech corporations scale securely and effectively on cloud.
 Christopher Brown, PhD, is head of the Discovery crew at Metagenomi. He’s an completed scientist and professional in metagenomics, and has led the invention and characterization of quite a few novel enzyme methods for gene enhancing functions.
Christopher Brown, PhD, is head of the Discovery crew at Metagenomi. He’s an completed scientist and professional in metagenomics, and has led the invention and characterization of quite a few novel enzyme methods for gene enhancing functions.
 Jamal Arif is a Senior Options Architect and Generative AI Specialist at AWS with over a decade of expertise serving to clients design and operationalize next-generation AI and cloud-native architectures. His work focuses on agentic AI, Kubernetes, and modernization frameworks, guiding enterprises by scalable adoption methods and production-ready design patterns. Jamal builds thought-leadership content material and speaks at AWS Summits, re:Invent, and business conferences, sharing finest practices for constructing safe, resilient, and high-impact AI options.
Jamal Arif is a Senior Options Architect and Generative AI Specialist at AWS with over a decade of expertise serving to clients design and operationalize next-generation AI and cloud-native architectures. His work focuses on agentic AI, Kubernetes, and modernization frameworks, guiding enterprises by scalable adoption methods and production-ready design patterns. Jamal builds thought-leadership content material and speaks at AWS Summits, re:Invent, and business conferences, sharing finest practices for constructing safe, resilient, and high-impact AI options.
 Pavel Novichkov, PhD, is a Senior Options Architect at AWS specializing in genomics and life sciences. He brings over 15 years of bioinformatics and cloud improvement expertise to assist healthcare and life sciences startups design and implement cloud-based options on AWS. He accomplished his postdoc on the Nationwide Heart for Biotechnology Data (NIH) and served as a Computational Analysis Scientist at Berkeley Lab for over 12 years, the place he co-developed revolutionary NGS-based know-how that was acknowledged amongst Berkeley Lab’s prime 90 breakthroughs in its historical past.
Pavel Novichkov, PhD, is a Senior Options Architect at AWS specializing in genomics and life sciences. He brings over 15 years of bioinformatics and cloud improvement expertise to assist healthcare and life sciences startups design and implement cloud-based options on AWS. He accomplished his postdoc on the Nationwide Heart for Biotechnology Data (NIH) and served as a Computational Analysis Scientist at Berkeley Lab for over 12 years, the place he co-developed revolutionary NGS-based know-how that was acknowledged amongst Berkeley Lab’s prime 90 breakthroughs in its historical past.