Make it work, then make it stunning, then should you actually, actually must, make it quick. 90 p.c of the time, should you make it stunning, it is going to already be quick. So actually, simply make it stunning! (Supply)
— Joe Armstrong (co-designers of the Erlang programming language.)
article about Python for the collection “Knowledge Science: From College to Work.” For the reason that starting, you’ve got realized how one can handle your Python undertaking with UV, how one can write a clear code utilizing PEP and SOLID rules, how one can deal with errors and use loguru to log your code and how one can write exams.
Now you might be ready to create working, production-ready code. However code is rarely good and might at all times be improved. A remaining (non-obligatory, however extremely beneficial) step in creating code is optimization.
To optimize your code, you want to have the ability to observe what’s happening in it. To take action, we use instruments referred to as Profilers. They generate profiles of your code. It means a set of statistics that describes how typically and for a way lengthy varied elements of this system executed. They make it doable to determine bottlenecks and elements of the code that devour too many sources. In different phrases, they present the place your code needs to be optimized.
Right now, there may be such a proliferation of profilers in Python that the default profiler in Pycharm is known as yappi for “But One other Python Profiler”.
This text is due to this fact not an exhaustive record of all current profilers. On this article, I current a device for every side of the code we need to profile: reminiscence, time and CPU/GPU consumption. Different packages can be talked about with some references however is not going to be detailed.
I – Reminiscence profilers
Reminiscence profiling is the strategy of monitoring and evaluating a program’s reminiscence utilization whereas working. This methodology helps builders to find reminiscence leaks, optimizing reminiscence utilization, and comprehending their packages’ reminiscence consumption patterns. Reminiscence profiling is essential to forestall purposes from utilizing extra reminiscence than needed and inflicting sluggish efficiency or crashes.
1/ memory-profiler
memory_profiler is an easy-to-use Python module designed to profile reminiscence utilization of a script. It relies on psutil module. To put in the bundle, merely kind:
pip set up memory_profiler # (in your digital surroundings)
# or should you use uv (what I encourage)
uv add memory_profilerProfiling executable
One of many benefits of this bundle is that it’s not restricted to pythonic use. It installs the mprof command that permits monitoring the exercise of any executable.
As an illustration, you possibly can monitor the reminiscence consummation of purposes like ollama by working this command:
mprof run ollama run gemma3:4b
# or with uv
mprof run ollama run gemma3:4bTo see the outcome, it’s a must to set up matplotlib first. Then, you possibly can plot the recorded reminiscence profile of your executable by working:
mprof plot
# or with uv
mprof run ollama run gemma3:4bThe graph then appears like this:

Profiling Python code
Let’s get again to what brings us right here, the monitoring of a Python code.
memory_profiler works with a line-by-line mode utilizing a easy decorator @profile. First, you enhance the curiosity operate and then you definitely run the script. The output can be written on to the terminal. Think about the next monitoring.py script:
@profile
def my_func():
a = [1] * (10 ** 6)
b = [2] * (2 * 10 ** 7)
del b
return a
if __name__ == '__main__':
my_func()It is very important discover that it’s not essential to import the bundle from memory_profiler import profile on the start of the script. On this case it’s a must to specify some particular arguments to the Python interpreter.
python-m memory_profiler monitoring.py # with an area between python and -m
# or
uv run -m memory_profiler monitoring.pyAnd you’ve got the next output with a line-by-line particulars:

The output is a desk with 5 columns.
- Line #: The road variety of the profiled code
- Mem utilization: The reminiscence utilization of the Python interpreter after executing that line.
- Increment: The change in reminiscence utilization in comparison with the earlier line.
- Occurrences: The variety of instances that line was executed.
- Line Contents: The precise supply code.
This output may be very detailed and permits very wonderful monitoring of a particular operate.
Vital: Sadly, this bundle is now not actively maintained. The creator is in search of a substitute.
2/ tracemalloc
tracemalloc is a built-in module in Python that tracks reminiscence allocations and deallocations. Tracemalloc offers an easy-to-use interface for capturing and analyzing reminiscence utilization snapshots, making it a useful device for any Python developer.
It provides the next particulars:
- Reveals the place every object was allotted by offering a traceback.
- Provides reminiscence allocation statistics by file and line quantity, together with the general measurement, rely, and common measurement of reminiscence blocks.
- Permits you to evaluate two snapshots to determine potential reminiscence leaks.
The bundle tracemalloc could also be usefull to determine reminiscence leak in your code.
Personally, I discover it much less intuitive to arrange than the opposite packages introduced on this article. Listed here are some hyperlinks to go additional:
II – Time profilers
Time profiling is the method of measuring the time spent in numerous elements of a program. By figuring out efficiency bottlenecks, you possibly can focus their optimization efforts on the elements of the code that can have essentially the most vital impression.
1/ line-profiler
The line-profiler bundle is sort of just like memory-profiler, however it serves a unique function. It’s designed to profile particular features by measuring the execution time of every line inside these features. To make use of LineProfiler successfully, you might want to explicitly specify which features you need it to profile by merely including the @profile decorator above them.
To put in it simply kind:
pip set up line_profiler # (in your digital surroundings)
# or
uv add line_profilerContemplating the next script named monitoring.py
@profile
def create_list(lst_len: int):
arr = []
for i in vary(0, lst_len):
arr.append(i)
def print_statement(idx: int):
if idx == 0:
print("Beginning array creation!")
elif idx == 1:
print("Array created efficiently!")
else:
increase ValueError("Invalid index supplied!")
@profile
def most important():
print_statement(0)
create_list(400000)
print_statement(1)
if __name__ == "__main__":
most important()To measure the execution time of the operate most important() and create_list(), we add the decorator @profile.
The best solution to get a time profiling of this script to make use of the kernprof script.
kernprof -lv monitoring.py # (in your digital surroundings)
# or
uv run kernprof -lv monitoring.pyIt’ll create a binary file named your_script.py.lprof. The argument -v permits to indicate directyl the output within the terminal.
In any other case, you possibly can view the outcomes later like so:
python-m line_profiler monitoring.py.lprof # (in your digital surroundings)
# or
uv run python -m line_profiler monitoring.py.lprofIt offers the next informations:

There are two tables, one by profiled operate. Every desk containes the next informations
- Line #: The road quantity within the file.
- Hits: The variety of instances that line was executed.
- Time: The overall period of time spent executing the road within the timer’s items. Within the header data earlier than the tables, you will note a line “Timer unit:” giving the conversion issue to seconds. It might be completely different on completely different methods.
- Per Hit: The typical period of time spent executing the road as soon as within the timer’s items
- % Time: The share of time spent on that line relative to the overall quantity of recorded time spent within the operate.
- Line Contents: The precise supply code.
1/ cProfile
Python comes with two built-in profilers:
cProfile: A C extension with affordable overhead that makes it appropriate for profiling long-running packages. It’s endorsed for many customers.profile: A pure Python module whose interface is imitated bycProfile, however which provides vital overhead to profiled packages. It may be a beneficial device when you might want to lengthen or customise the profiling performance.
The bottom syntax is cProfile.run(assertion, filename=None, kind=-1). The filename argument could be handed to avoid wasting the output. And the kind argument can be utilized to specify how the output needs to be printed. By default, it’s set to -1( no worth).
As an illustration, should you modify the monitoring script like this:
import cProfile
def create_list(lst_len: int):
arr = []
for i in vary(0, lst_len):
arr.append(i)
def print_statement(idx: int):
if idx == 0:
print("Beginning array creation!")
elif idx == 1:
print("Array created efficiently!")
else:
increase ValueError("Invalid index supplied!")
def most important():
print_statement(0)
create_list(400000)
print_statement(1)
if __name__ == "__main__":
cProfile.run("most important()")
we’ve got the next output:

First, we’ve got the script outputs: print_statement(0) and print_statement(1).
Then, we’ve got the profiler output: The primary line reveals the variety of operate calls and the time it took to run. The second line is a reminder of the sorted parameter. And, the profiler offers a desk with six columns:
- ncalls: Reveals the variety of calls made
- tottime: Complete time taken by the given operate. Word that the time made in calls to sub-functions are excluded.
- percall: Complete time / No of calls. (the rest is omitted)
- cumtime: Not like tottime, this consists of time spent on this and all subfunctions that the higher-level operate calls. It’s most helpful and is correct for recursive features.
- percall: The percall following cumtime is calculated because the quotient of cumtime divided by primitive calls. The primitive calls embrace all of the calls that weren’t included via recursion.
- filename: The title of the tactic.
The primary and the final rows of the desk come from cProfile. The opposite rows are concerning the script.
You possibly can customise the output by utilizing the Profile() class. First, it’s a must to initialize an occasion of Profile class and utilizing the tactic allow() and disable() to, respectively, begin and to finish the amassing of profiling knowledge. Then, the pstats module can be utilized to govern the outcomes collected by the profiler object.
To kind output by cumulative time, as an alternative of the usual title the earlier code could be rewritten like this:
import cProfile, pstats
# ...
# Identical as earlier than
if __name__ == "__main__":
profiler = cProfile.Profile()
profiler.allow()
most important()
profiler.disable()
stats = pstats.Stats(profiler).sort_stats('cumtime')
stats.print_stats()
And the output turns into:

As you possibly can see, now the desk is sorted by cumtime. And the 2 rows of cProfile of the earlier desk should not on this desk.
Visualize profiling with Snakeviz.
The output may be very simple to analyse. However, it could possibly develop into unreadable if the profiled code turns into too large.
One other solution to analyse the ouput is to visualise knowledge as an alternative of learn it. To take action, we use the Snakeviz bundle. To put in it, merely kind:
pip set up snakeviz # (in your digital surroundings)
# or
uv add snakevizThen, substitute stats.print_stats() by stats.dump_stats("profile.prof") to avoid wasting profiling knowledge. Now, you possibly can have a visualization of your profiling by typing:
snakeviz profile.profIt launches a file browser interface from which you’ll select amongst two knowledge visualizations: Icicle and Sunburst.


It’s simpler to learn than the print_stats() output as a result of you possibly can work together with every ingredient by shifting your mouse over it. As an illustration, you possibly can have extra particulars concerning the operate create_list()

evaluate_model() (from the creator).Create a name graph with gprof2dot
A name graph is a visible illustration of the relationships between features or strategies in a program, displaying which features name others and the way lengthy every operate or methodology takes. It may be seen as a map of your code.
pip set up gprof2dot # (in your digital surroundings)
# or
uv add gprof2dotThen exectute your by typing
python-m cProfile -o monitoring.pstats .monitoring.py # (in your digital surroundings)
# or
uv run python-m cProfile -o monitoring.pstats .monitoring.py
It’ll create a monitoring.pstats that may be flip right into a name graph utilizing the next command:
gprof2dot -f pstats monitoring.pstats | dot -Tpng -o monitoring.png # (in your digital surroundings)
# or
uv run gprof2dot -f pstats monitoring.pstats | dot -Tpng -o monitoring.pngThen the decision graph is saved right into a png file named monitoring.png

2/ Different attention-grabbing packages
a/ PyCallGraph
PyCallGraph is a Python module that creates name graph visualizations. To make use of it, it’s a must to :
To create a name graph of your code, provide run it a PyCallGraph context like this:
from pycallgraph import PyCallGraph
from pycallgraph.output import GraphvizOutput
with PyCallGraph(output=GraphvizOutput()):
# code you need to profileThen, you get a png of the decision graph of your code is called by default pycallgraph.png.
I’ve made the decision graph of the earlier instance:

In every field, you’ve got the title of the operate, the time spent in and the variety of calls. Like with snakeviz, the graph could also be very complicated in case your code has many dependencies. However the shade signifies the bottlenecks. In complicated code, it’s very attention-grabbing to check it to see the dependencies and relationships.
b/ PyInstrument
PyInstrument can be a Python profiler very simple to make use of. You possibly can add the profiler in your script by surredning the code like this:
from pyinstrument import Profiler
profiler = Profiler()
profiler.begin()
# code you need to profile
profiler.cease()
print(profiler.output_text(unicode=True, shade=True))The output provides

It’s much less detailled than cProfile however it’s also extra readable. Your features are highlighted and sorted by time.
Butthe true curiosity of PyInstrument comes with its html output. To get this html output merely kind within the terminal:
pyinstrument --html .monitoring.py
# or
uv run pyinstrument --html .monitoring.pyIt launches a file browser interface from which you’ll select amongst two knowledge visualizations: Name stack and Timeline.


Right here, the profile is extra detailed and you’ve got many choices to filter.
CPU/GPU profiler
CPU and GPU profiling is the method of analyzing the utilization and efficiency of a program on the central processing unit (CPU) and graphics processing unit (GPU). By measuring how a lot sources are spent on completely different elements of the code on these processing items, builders can determine efficiency bottlenecks, perceive the place their code is being executed, and optimize their software to realize higher efficiency and effectivity.
So far as I do know, there is just one bundle that may profile GPU energy consumption.
1/ Scalene
Scalene is a high-performance CPU, GPU and reminiscence profiler designed particularly for Python. It’s an open-source bundle that gives detailed insights. It’s designed to be quick, correct, and straightforward to make use of, making it a wonderful device for builders trying to optimize their code.
- CPU/GPU Profiling: Scalene offers detailed data on CPU/GPU utilization, together with the time spent in numerous elements of your code. It may possibly assist you to determine efficiency bottlenecks and optimize your code for higher execution instances.
- Reminiscence Profiling: Scalene tracks reminiscence allocation and deallocation, serving to you perceive how your code makes use of reminiscence. That is significantly helpful for figuring out reminiscence leaks or optimizing memory-intensive purposes.
- Line-by-Line Profiling: Scalene offers line-by-line profiling, which provides you an in depth breakdown of the time spent in every line of your code. This function is invaluable for pinpointing efficiency points.
- Visualization: Scalene features a graphical interface for visualizing profiling outcomes, making it simpler to grasp and navigate the info.
To spotlight all some great benefits of Scalene, I’ve developed features with the only real intention of consuming reminiscence memory_waster(), CPU cpu_waster() and GPU gpu_convolution(). All of them are in a script scalene_tuto.py.
import random
import copy
import math
import cupy as cp
import numpy as np
def memory_waster():
"""Wastes reminiscence however in a managed means"""
memory_hogs = []
# Create reasonably sized redundant knowledge constructions
for i in vary(100):
garbage_data = []
for j in vary(1000):
waste = f"Ineffective string #{j} repeated " * 10
garbage_data.append(waste)
garbage_data.append(
{
"id": j,
"knowledge": waste,
"numbers": [random.random() for _ in range(50)],
"range_data": record(vary(100)),
}
)
memory_hogs.append(garbage_data)
for iteration in vary(4):
print(f"Creating copy #{iteration}...")
memory_copy = copy.deepcopy(memory_hogs)
memory_hogs.lengthen(memory_copy)
return memory_hogs
def cpu_waster():
meaningless_result = 0
for i in vary(10000):
for j in vary(10000):
temp = (i**2 + j**2) * random.random()
temp = temp / (random.random() + 0.01)
temp = abs(temp**0.5)
meaningless_result += temp
# Some trigonometric operations
angle = random.random() * math.pi
temp += math.sin(angle) * math.cos(angle)
if i % 100 == 0:
random_mess = [random.randint(1, 1000) for _ in range(1000)] # Smaller record
random_mess.kind()
random_mess.reverse()
random_mess.kind()
return meaningless_result
def gpu_convolution():
image_size = 128
kernel_size = 64
picture = np.random.random((image_size, image_size)).astype(np.float32)
kernel = np.random.random((kernel_size, kernel_size)).astype(np.float32)
image_gpu = cp.asarray(picture)
kernel_gpu = cp.asarray(kernel)
outcome = cp.zeros_like(image_gpu)
for y in vary(kernel_size // 2, image_size - kernel_size // 2):
for x in vary(kernel_size // 2, image_size - kernel_size // 2):
pixel_value = 0
for ky in vary(kernel_size):
for kx in vary(kernel_size):
iy = y + ky - kernel_size // 2
ix = x + kx - kernel_size // 2
pixel_value += image_gpu[iy, ix] * kernel_gpu[ky, kx]
outcome[y, x] = pixel_value
result_cpu = cp.asnumpy(outcome)
cp.cuda.Stream.null.synchronize()
return result_cpu
def most important():
print("n1/ Losing some reminiscence (managed)...")
_ = memory_waster()
print("n2/ Losing CPU cycles (managed)...")
_ = cpu_waster()
print("n3/ Losing GPU cycles (managed)...")
_ = gpu_convolution()
if __name__ == "__main__":
most important()For the GPU operate, it’s a must to set up cupy in response to your cuda model (nvcc --version to get it)
pip set up cupy-cuda12x # (in your digital surroundings)
# or
uv add set up cupy-cuda12xAdditional particulars on putting in cupy could be discovered within the documentation.
To run Scalene, use the command
scalene scalene_tuto.py
# or
uv run scalene scalene_tuto.pyIt profiles each CPU, GPU, and reminiscence by default. In case you solely need one or among the choices, use the flags --cpu, --gpu, and --memory.
Scalene offers a line-level and a operate stage profiling. And it has two interfaces: the Command Line Interface (CLI) and the online interface.
Vital: It’s higher to make use of Scalene with Ubuntu utilizing WSL. In any other case, the profiler doesn’t retrieve reminiscence consumption data.
a) Command Line Interface
By default, Scalene’s output is the online interface. To acquire the CLI as an alternative, add the flag --cli.
scalene scalene_tuto.py --cli
# or
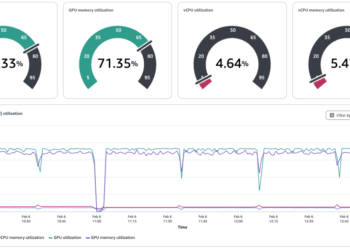
uv run scalene scalene_tuto.py --cliYou’ve the next outcomes:


By default, the code is displayed in darkish mode. So if, like me, you’re employed in gentle mode, the outcome isn’t very fairly.
The visualization is categorized into three distinct colours, every representing a unique profiling metric.
- The blue part represents CPU profiling, which offers a breakdown of the time spent executing Python code, native code (similar to C or C++), and system-related duties (like I/O operations).
- The inexperienced part is devoted to reminiscence profiling, displaying the proportion of reminiscence allotted by Python code, in addition to the general reminiscence utilization over time and its peak values.
- The yellow part focuses on GPU profiling, displaying the GPU’s working time and the quantity of information copied between the GPU and CPU, measured in mb/s. It’s value noting that GPU profiling is presently restricted to NVIDIA GPUs.
b) The online interface.
The online interface is split in three elements.



The colour code is similar as within the command lien interface. However some icons are added:
- 💥: Optimizable code area (efficiency indication within the Operate Profile part).
- ⚡: Optimizable traces of code.
c) AI Ideas
One of many nice benefits of Scalene is the power to make use of AI to enhance the slowness and/or overconsumption you’ve got recognized. It presently helps OpenAI API, Amazon BedRock, Azure OpenAI and ollama in native

After choosing your instruments, you simply must click on on 💥 or ⚡if you wish to optimize part of the code or only a line.
I take a look at it with codellama:7b-python from ollama to optimize the gpu_convolution() operate. Sadly, as talked about within the interface:
Word that optimizations are AI-generated and will not be appropriate.
Not one of the instructed optimizations labored. However the codebase was not conducive to optimization because it was artificially difficult. Simply take away pointless traces to avoid wasting time and reminiscence. Additionally, I used a small mannequin, which might be the explanation.
Despite the fact that my exams had been inconclusive, I feel this selection could be attention-grabbing and can absolutely proceed to enhance.
Conclusion
These days, we’re much less involved concerning the useful resource consumption of our developments, and really rapidly these optimization deficits can accumulate, making the code gradual, too gradual for manufacturing, and generally even requiring the acquisition of extra highly effective {hardware}.
Code profiling instruments are indispensable in the case of figuring out areas in want of optimization.
The mix of the reminiscence profiler and line profiler offers an excellent preliminary evaluation: simple to arrange, with easy-to-understand studies.
Instruments similar to cProfile and Scalene are full and have graphical representations, however require extra time to investigate. Lastly, the AI optimization choice provided by Scalene is an actual asset, even when in my case the mannequin used was not ample to offer something related.
Interested by Python & Knowledge Science?
Comply with me for extra tutorials and insights!