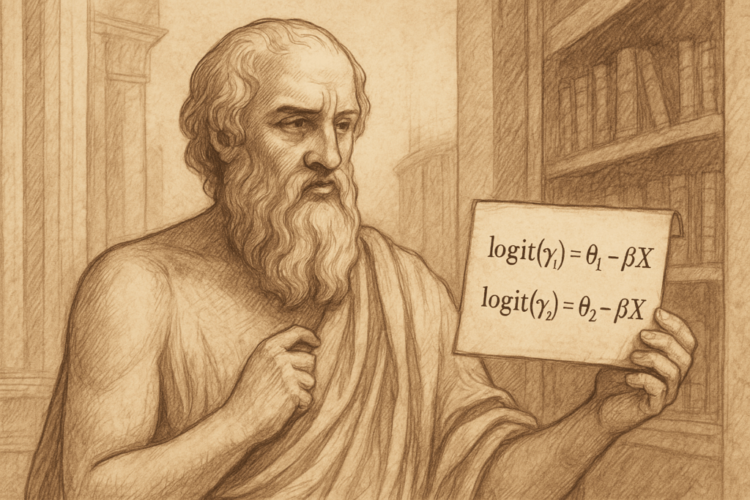You could find the total code for this instance on the backside of this submit.
odds mannequin for ordinal logistic regression was first launched by McCullagh (1980). This mannequin extends binary logistic regression to conditions the place the dependent variable is ordinal [it consists of ordered categorical values]. The proportional odds mannequin is constructed on a number of assumptions, together with independence of observations, linearity of the log-odds, absence of multicolinearity amongst predictors, and the proportional odds assumption. This final assumption states that the regression coefficients are fixed throughout all thresholds of the ordinal dependent variable. Making certain the proportional odds assumption holds is essential for the validity and interpretability of the mannequin.
Quite a lot of strategies have been proposed within the literature to evaluate mannequin match and, specifically, to check the proportional odds assumption. On this paper, we deal with two approaches developed by Brant in his article Brant (1990), “Assessing Proportionality within the Proportional Odds Mannequin for Ordinal Logistic Regression.” We additionally reveal tips on how to implement these strategies in Python, making use of them to real-world information. Whether or not you come from a background in information science, machine studying, or statistics, this text goals to assist your perceive tips on how to consider mannequin slot in ordinal logistic regression.
This paper is organized into 4 most important sections:
- The primary part introduces the proportional odds mannequin and its assumptions.
- The second part discusses tips on how to assess the proportional odds assumption utilizing the probability ratio check.
- The third part covers the evaluation of the proportional odds assumption utilizing the separate suits strategy.
- The ultimate part gives examples, illustrating the implementation of those evaluation strategies in Python with information.
1. Introduction to the Proportional Odds Mannequin
Earlier than presenting the mannequin, we introduce the information construction. We assume we’ve N unbiased observations. Every commentary is represented by a vector of p explanatory variables Xi = (Xi1, Xi2, …, Xip), together with a dependent or response variable Y that takes ordinal values from 1 to Okay. The proportional odds mannequin particularly fashions the cumulative distribution chances of the response variable Y, outlined as γj = P(Y ≤ j | Xi) for j = 1, 2, …, Okay – 1, as capabilities of the explanatory variables Xi. The mannequin is formulated as follows:
logit(γⱼ) = log(γⱼ / (1 − γⱼ)) = θⱼ − βᵀ𝐗. (1)
The place θⱼ are the intercepts for every class j and respect the situation θ₁ < θ₂ < ⋯ < θₖ₋₁, and β is the vector of regression coefficients that are the identical for all classes.
We observe a monotonic development within the coefficients θⱼ throughout the classes of the response variable Y.
This mannequin is also referred to as the grouped steady mannequin, as it may be derived by assuming the existence of a steady latent variable Y∗. This latent variable follows a linear regression mannequin with conditional imply η = βᵀ𝐗, and it pertains to the noticed ordinal variable Y by thresholds θⱼ outlined as follows:
y∗ = βᵀ𝐗+ϵ, (2)
the place ϵ is an error time period (random noise), usually assumed to comply with a typical logistic distribution within the proportional odds mannequin.
The latent variable Y* is unobserved and partitioned into intervals outlined by thresholds θ₁, θ₂, …, θₖ₋₁, producing the noticed ordinal variable Y as follows:

Within the subsequent part, we introduce the assorted approaches proposed by Brant (1990) for assessing the proportional odds assumption. These strategies consider whether or not the regression coefficients stay fixed throughout the classes outlined by the ordinal response variable.
2. Assessing the Proportional Odds Assumption: The Probability Ratio Take a look at
To evaluate the proportional odds assumption in an ordinal logistic regression mannequin, Brant (1990) proposes the usage of the probability ratio check. This strategy begins by becoming a much less restrictive mannequin through which the regression coefficients are allowed to fluctuate throughout classes. This mannequin is expressed as:
logit(γj) = θj − βjT𝐗. for j = 1, …, Okay-1 (4)
the place βj is the vector [vector of dimension p] of regression coefficients for every class j. Right here the coefficients βj are allowed to fluctuate throughout classes, which implies that the proportional odds assumption just isn’t glad. We then use the conventionnel probability ratio check to evaluate the speculation :
H0 : βj = β for all j = 1, 2, …, Okay−1. (5)
To carry out this check, we conduct a probability ratio check evaluating the unconstrained (non-proportional or satured) mannequin with the constrained (proportional odds or lowered) mannequin.
Earlier than continuing additional, we briefly recall tips on how to use the probability ratio check in speculation testing. Suppose we wish to consider the null speculation H0 : θ ∈ Θ0 in opposition to the choice H1 : θ ∈ Θ1,
The probability ratio statistic is outlined as:

the place 𝓛(θ) is the probability operate, θ̂ is the utmost probability estimate (MLE) below the total mannequin, and θ̂₀ is the MLE below the constrained mannequin. The check statistic λ follows a chi-square distribution with levels of freedom equal to the distinction within the variety of parameters between the total and constrained fashions.
Right here, θ̂ is the utmost probability estimate (MLE) below the total (unconstrained) mannequin, and θ̂₀ is the MLE below the constrained mannequin the place the proportional odds assumption holds. The
check statistic λ follows a chi-square distribution below the null speculation.
In a basic setting, suppose the total parameter house is denoted by
Θ = (θ₁, θ₂, …, θq, …, θp),
and the restricted parameter house below the null speculation is
Θ₀ = (θ₁, θ₂, …, θq).
(Notice: These parameters are generic and shouldn’t be confused with the Okay−1 thresholds or intercepts within the proportional odds mannequin.), the probability ratio check statistic λ follows a chi-square distribution with p−q levels of freedom. The place p represents the overall variety of parameters within the full (unconstrained or “saturated”) mannequin, whereas Okay−1 corresponds to the variety of parameters within the lowered (restricted) mannequin.
Now, allow us to apply this strategy to the ordinal logistic regression mannequin with the proportional odds assumption. Assume that our response variable has Okay ordered classes and that we’ve p predictor variables. To make use of the probability ratio check to guage the proportional odds assumption, we have to examine two fashions:
1. Unconstrained Mannequin (non-proportional odds):
This mannequin permits every consequence threshold to have its personal set of regression coefficients, that means that we don’t assume the regression coefficients are equal throughout all thresholds. The mannequin is outlined as:
logit(γj) = θj − βjT𝐗. for j = 1, …, Okay-1 (7)
- There are Okay−1 threshold (intercept) parameters: θ1, θ2, … , θOkay-1
- Every threshold has its personal vector of slope coefficients βj of dimension p
Thus, the overall variety of parameters within the unconstrained mannequin is:
(Okay−1) thresholds + (Okay−1)×p slopes = (Okay−1)(p+1).
2. Proportional Odds Mannequin:
This mannequin assumes a single set of regression coefficients for all thresholds:
logit(γj) = θj − βT𝐗. for j = 1, …, Okay-1 (8)
- There are Okay−1 threshold parameters
- There’s one frequent slope vector β (of dimension p x 1) for all j
Thus, the overall variety of parameters within the proportional odds mannequin is:
(Okay−1) thresholds+p slopes=(Okay−1)+p
Thus, the probability ratio check statistic follows a chi-square distribution with levels of freedom:
df = [(K−1)×(p+1)]−[(K−1)+p] = (Okay−2)×p
This check gives a proper approach to assess whether or not the proportional odds assumption holds for the given information. At a significance stage of 1%, 5%, or every other standard threshold, the proportional odds assumption is rejected if the check statistic λ exceeds the crucial worth from the chi-square distribution with (Okay−2)×p levels of freedom.
In different phrases, we reject the null speculation
H0 : β1 = β2 = ⋯ =βOkay-1 = β,
which states that the regression coefficients are equal throughout all cumulative logits. This check has the benefit of being simple to implement and gives an general evaluation of the proportional odds assumption.
Within the subsequent part, we introduce the proportional odds check primarily based on separate suits.
3. Assessing the Proportional Odds Assumption: The Separate Matches Method.
To grasp this part, it’s essential first to grasp the idea and properties of the Mahalanobis distance. This distance is used to quantify how completely different two vectors are in a multivariate house that shares the identical distribution.
Let x = (x₁, x₂, …, xₚ)ᵀ and y = (y₁, y₂, …, yₚ)ᵀ. The Mahalanobis distance between them is outlined as:

the place Σ is the covariance matrix of the distribution. The Mahalanobis distance squared is carefully associated to the chi-squared (χ²) distribution. Particularly, if X ∼ N(μ, Σ) is a p-dimensional usually distributed random vector with imply μ and covariance matrix Σ, then the Mahalanobis distance Dₘ(X, μ)2 follows a χ² distribution with p levels of freedom.
Understanding this relationship is essential for evaluating proportionality utilizing separate mannequin suits — a subject we’ll return to shortly.
The truth is, the creator notes that the pure strategy to evaluating the proportional odds assumption is to suit a set of Okay−1 binary logistic regression fashions (the place Okay is the variety of classes of the response variable), after which use the statistical properties of the estimated parameters to assemble a check statistic for the proportional odds speculation.
The process is as follows:
First, we assemble separate binary logistic regression fashions for every threshold j = 1, 2, …, Okay−1 of the ordinal response variable Y. For every threshold j, we outline a binary variable Zj, which takes the worth 1 if the commentary exceeds threshold j, and 0 in any other case. Particularly, we’ve:

With the chance, πj = P(Zj = 1 | X) = 1−γj satisfying the logistic regression mannequin:
logit(πj ) = θj − βT𝐗. for j = 1, …, Okay-1 (10)
Then, assessing the proportional odds assumption on this context includes testing the speculation that the regression coefficients βj are equal throughout all Okay−1 fashions. That is equal to testing the speculation:
H0 : β1 = β2 = ⋯ = βOkay-1 (11)
Let β̂ⱼ denote the utmost probability estimators of the regression coefficients for every binary mannequin, and let β̂ = (β̂₁ᵀ, β̂₂ᵀ, …, β̂ₖ₋₁ᵀ)ᵀ signify the worldwide vector of estimators. This vector is asymptotically usually distributed, such that 𝔼(β̂ⱼ) ≈ β, with variance-covariance matrix 𝕍(β̂ⱼ). The final time period of this matrix, cov(β̂ⱼ, β̂ₖ), must be decided and is given by:

the place Cov(β̂ⱼ, β̂ₖ) is the covariance between the estimated coefficients of the j-th and k-th binary fashions. The asymptotic covariances, Cov(β̂ⱼ, β̂ₖ), are obtained by deleting the primary row and column of:
(X+T Wjj X₊)-1X+TWjlX+ (Xᵗ WllX₊)-1
the place Wjl: n × n is diagonal with typical entry πl − πj πl for j ≤ l, and X+: n x p matrix denotes the matrix of explanatory variables augmented on the left with a column of ones [1’s].
To guage the proportional odds assumption, Brant constructs a matrix 𝐃 that captures the variations between the coefficients β̂ⱼ. Recall that every vector β̂ⱼ has dimension p. The matrix 𝐃 is outlined as follows:

the place 𝐈 is the identification matrix of measurement p × p. The primary row of the matrix 𝐃 corresponds to the distinction between the primary and second coefficients, the second row corresponds to the distinction between the second and third coefficients, and so forth, till the final row which corresponds to the distinction between the (Okay − 2)-th and (Okay − 1)-th coefficients. We will discover that the product 𝐃β̂ will yield a vector of variations between the coefficients β̂ⱼ.
As soon as the matrix 𝐃 is constructed, Brant defines the Wald statistic X² to check the proportional odds assumption. This statistic may be interpreted because the Mahalanobis distance between the vector 𝐃β̂ and the zero vector. The Wald statistic is outlined as follows:

which shall be asymptotically χ² distributed with (Okay − 2)p levels of freedom below the null speculation. The difficult half right here is to find out the variance-covariance matrix 𝕍̂(β̂). In his article, Brant gives an specific estimator for this variance-covariance matrix, which is predicated on the utmost probability estimators β̂ⱼ from every binary mannequin.
Within the following sections, we implement these approaches in Python, utilizing the statsmodels package deal for the regressions and statistical assessments.
Instance: Utility of the Two Proportional Odds Exams
The info for this instance comes from the “Wine High quality” dataset, which accommodates details about pink wine samples and their high quality scores. The dataset contains 1,599 observations and 12 variables. The goal variable, “high quality,” is ordinal and initially ranges from 3 to eight. To make sure sufficient observations in every group, we mix classes 3 and 4 right into a single class (labeled 4), and classes 7 and eight right into a single class (labeled 7), so the response variable has 4 ranges. We then deal with outliers within the explanatory variables utilizing the Interquartile Vary (IQR) methodology. Lastly, we choose three predictors—unstable acidity, free sulfur dioxide, and whole sulfur dioxide—to make use of in our ordinal logistic regression mannequin, and we standardize these variables to have a imply of 0 and a typical deviation of 1.
Tables 1 and a couple of under current the outcomes of the three binary logistic regression fashions and the proportional odds mannequin, respectively. A number of discrepancies may be seen in these tables, significantly within the “unstable acidity” coefficients. For example, the distinction within the “unstable acidity” coefficient between the primary and second binary fashions is -0.280, whereas the distinction between the second and third fashions is 0.361. These variations recommend that the proportional odds assumption might not maintain [I also suggest to compute standard errors of difference for more confidence].


To evaluate the general significance of the proportional odds assumption, we carry out the probability ratio check, which yields a check statistic of LR = 53.207 and a p-value of 1.066 × 10-9 when in comparison with the chi-square distribution with 6 levels of freedom. This consequence signifies that the proportional odds assumption is violated on the 5% significance stage, suggesting that the mannequin might not be applicable for the information. We additionally use the separate suits strategy to additional examine this assumption. The Wald check statistic is computed as X2 = 41.880, with a p-value of 1.232 × 10-7, additionally primarily based on the chi-square distribution with 6 levels of freedom. This additional confirms that the proportional odds assumption is violated on the 5% significance stage.
Conclusion
This paper had two most important targets: first, for instance tips on how to check the proportional odds assumption within the context of ordinal logistic regression, and second, to encourage readers to discover Brant (1990)’s article for a deeper understanding of the subject.
Brant’s work extends past assessing the proportional odds assumption—it additionally gives strategies for evaluating the general adequacy of the ordinal logistic regression mannequin. For example, he discusses tips on how to check whether or not the latent variable Y∗ actually follows a logistic distribution or whether or not an alternate hyperlink operate is likely to be extra applicable.
On this article, we centered on a worldwide evaluation of the proportional odds assumption, with out investigating which particular coefficients could also be liable for any violations. Brant additionally addresses this finer-grained evaluation, which is why we strongly encourage you to learn his article in full.
Picture Credit
All photographs and visualizations on this article had been created by the creator utilizing Python (pandas, matplotlib, seaborn, and plotly) and excel, except in any other case acknowledged.
References
Brant, Rollin. 1990. “Assessing Proportionality within the Proportional Odds Mannequin for Ordinal Logistic Regression.” Biometrics, 1171–78.
McCullagh, Peter. 1980. “Regression Fashions for Ordinal Information.” Journal of the Royal Statistical Society: Sequence B (Methodological) 42 (2): 109–27.
Wasserman, L. (2013). All of statistics: a concise course in statistical inference. Springer Science & Enterprise Media.
Cortez, P., Cerdeira, A., Almeida, F., Matos, T., & Reis, J. (2009). Wine High quality [Dataset]. UCI Machine Studying Repository. https://doi.org/10.24432/C56S3T.
Information & Licensing
The dataset used on this article is licensed below the Inventive Commons Attribution 4.0 Worldwide (CC BY 4.0) license.
It permits to be used, distribution, and adaptation, even for business functions, offered that applicable credit score is given.
The unique dataset was printed by [Name of the Author / Institution] and is out there right here.
Codes
Import information and the variety of observations
import pandas as pd
information = pd.read_csv("winequality-red.csv", sep=";")
information.head()
# Repartition de la variable cible high quality
information['quality'].value_counts(normalize=False).sort_index()
# I wish to regroup modalities 3, 4 and the modalities 7 and eight
information['quality'] = information['quality'].exchange({3: 4, 8: 7})
information['quality'].value_counts(normalize=False).sort_index()
print("Variety of observations:", information.form[0])Let’s deal with the outliers of the predictor variables (excluding the goal variable high quality) utilizing the IQR methodology.
def remove_outliers_iqr(df, column):
Q1 = df[column].quantile(0.25)
Q3 = df[column].quantile(0.75)
IQR = Q3 - Q1
lower_bound = Q1 - 1.5 * IQR
upper_bound = Q3 + 1.5 * IQR
return df[(df[column] >= lower_bound) & (df[column] <= upper_bound)]
for col in information.columns:
if col != 'high quality':
information = remove_outliers_iqr(information, col)Create a boxplot of every variable per group of high quality
var_names_without_quality = [col for col in data.columns if col != 'quality']
## Create the boxplot of every variable per group of high quality
import matplotlib.pyplot as plt
import seaborn as sns
plt.determine(figsize=(15, 10))
for i, var in enumerate(var_names_without_quality):
plt.subplot(3, 4, i + 1)
sns.boxplot(x='high quality', y=var, information=information)
plt.title(f'Boxplot of {var} by high quality')
plt.xlabel('High quality')
plt.ylabel(var)
plt.tight_layout()
plt.present()Implement Ordinal regression for the odd proportionality check.
# Implement the ordered logistic regression to variables 'unstable acidity', 'free sulfur dioxide', and 'whole sulfur dioxide'
from statsmodels.miscmodels.ordinal_model import OrderedModel
from sklearn.preprocessing import StandardScaler
explanatory_vars = ['volatile acidity', 'free sulfur dioxide', 'total sulfur dioxide']
# Standardize the explanatory variables
information[explanatory_vars] = StandardScaler().fit_transform(information[explanatory_vars])
def fit_ordered_logistic_regression(information, response_var, explanatory_vars):
mannequin = OrderedModel(
information[response_var],
information[explanatory_vars],
distr='logit'
)
consequence = mannequin.match(methodology='bfgs', disp=False)
return consequence
response_var = 'high quality'
consequence = fit_ordered_logistic_regression(information, response_var, explanatory_vars)
print(consequence.abstract())
# Compute the log-likelihood of the mannequin
log_reduced = consequence.llf
print(f"Log-likelihood of the lowered mannequin: {log_reduced}")Compute the total multinomial mannequin
# The probability ratio check
# Compute the total multinomial mannequin
import statsmodels.api as sm
data_sm = sm.add_constant(information[explanatory_vars])
model_full = sm.MNLogit(information[response_var], data_sm)
result_full = model_full.match(methodology='bfgs', disp=False)
#abstract
print(result_full.abstract())
# Commpute the log-likelihood of the total mannequin
log_full = result_full.llf
print(f"Log-likelihood of the total mannequin: {log_full}")
# Compute the probability ratio statistic
LR_statistic = 2 * (log_full - log_reduced)
print(f"Probability Ratio Statistic: {LR_statistic}")
# Compute the levels of freedom
df1 = (num_of_thresholds - 1) * len(explanatory_vars)
df2 = result_full.df_model - OrderedModel(
information[response_var],
information[explanatory_vars],
distr='logit'
).match().df_model
print(f"Levels of Freedom: {df1}")
print(f"Levels of Freedom for the total mannequin: {df2}")
# Compute the p-value
from scipy.stats import chi2
print("The LR statistic :", LR_statistic)
p_value = chi2.sf(LR_statistic, df1)
print(f"P-value: {p_value}")
if p_value < 0.05:
print("Reject the null speculation: The proportional odds assumption is violated.")
else:
print("Fail to reject the null speculation: The proportional odds assumption holds.")The code under constructs the Wald statistic X² = (𝐃β̂)ᵀ [𝐃𝕍̂(β̂)𝐃ᵀ]⁻¹ (𝐃β̂)
Okay-1 Binary Logit Estimation for Proportional Odds Assumption Test
import numpy as np
import statsmodels.api as sm
import pandas as pd
def fit_binary_models(information, explanatory_vars, y):
"""
Matches a sequence of binary logistic regression fashions to evaluate the proportional odds assumption.
Parameters:
- information: Authentic pandas DataFrame (should embody all variables)
- explanatory_vars: Listing of predictor variables (options)
- y: Array-like ordinal goal variable (n,) — e.g., classes like 4, 5, 6, 7
Returns:
- binary_models: Listing of fitted binary Logit mannequin objects (statsmodels)
- beta_hat: (Okay−1) × (p+1) array of estimated coefficients (together with intercept)
- var_hat: Listing of (p+1) × (p+1) variance-covariance matrices for every mannequin
- z_mat: DataFrame containing the binary reworked targets z_j (for inspection/debugging)
- thresholds: Listing of thresholds used to create the binary fashions
"""
qualities = np.type(np.distinctive(y)) # Sorted distinctive classes of y
thresholds = qualities[:-1] # Thresholds to outline binary fashions (Okay−1)
p = len(explanatory_vars)
n = len(y)
K_1 = len(thresholds)
binary_models = []
beta_hat = np.full((K_1, p+1), np.nan) # To retailer estimated coefficients
p_values_beta_hat = np.full((K_1, p+1), np.nan) # To retailer p-values
var_hat = []
z_mat = pd.DataFrame(index=np.arange(n)) # For storing binary goal variations
X_with_const = sm.add_constant(information[explanatory_vars]) # Design matrix with intercept
# Match one binary logistic regression for every threshold
for j, t in enumerate(thresholds):
z_j = (y > t).astype(int) # Binary goal: 1 if y > t, else 0
z_mat[f'z>{t}'] = z_j
mannequin = sm.Logit(z_j, X_with_const)
res = mannequin.match(disp=0)
binary_models.append(res)
beta_hat[j, :] = res.params.values # Retailer coefficients (with intercept)
p_values_beta_hat[j, :] = res.pvalues.values
var_hat.append(res.cov_params().values) # Retailer full (p+1) × (p+1) covariance matrix
return binary_models, beta_hat, X_with_const, var_hat, z_mat, thresholds
# Apply the operate to the information
binary_models, beta_hat, X_with_const, var_hat, z_mat, thresholds = fit_binary_models(
information, explanatory_vars, information[response_var]
)
# Show the estimated coefficients
print("Estimated coefficients (beta_hat):")
print(beta_hat)
# Show the design matrix (predictors with intercept)
print("Design matrix X (with fixed):")
print(X_with_const)
# Show the thresholds used to outline the binary fashions
print("Thresholds:")
print(thresholds)
# Show first few rows of the binary response matrix
print("z_mat (created binary goal variables):")
print(z_mat.head())
Compute Fitted Possibilities (π̂) for establishing the Wjl n x n diagonal matrix.
def compute_pi_hat(binary_models, X_with_const):
"""
Computes the fitted chances π̂ for every binary logistic regression mannequin.
Parameters:
- binary_models: Listing of fitted Logit mannequin outcomes (from statsmodels)
- X_with_const : 2D array of form (n, p+1) — the design matrix with intercept included
Returns:
- pi_hat: 2D array of form (n, Okay−1) containing the anticipated chances
from every of the Okay−1 binary fashions
"""
n = X_with_const.form[0] # Variety of observations
K_1 = len(binary_models) # Variety of binary fashions (Okay−1)
pi_hat = np.full((n, K_1), np.nan) # Initialize prediction matrix
# Compute fitted chances for every binary mannequin
for m, mannequin in enumerate(binary_models):
pi_hat[:, m] = mannequin.predict(X_with_const)
return pi_hat
# Assuming you have already got:
# - binary_models: checklist of fitted fashions from earlier operate
# - X_with_const: design matrix with intercept (n, p+1)
pi_hat = compute_pi_hat(binary_models, X_with_const)
# Show the form and a preview of the anticipated chances matrix
print("Form of pi_hat:", pi_hat.form) # Anticipated form: (n, Okay−1)
print("Preview of pi_hat:n", pi_hat[:5, :]) # First 5 rows
Compute the general estimated covariance matrix V̂(β̃) in two steps.
import numpy as np
# Assemble World Variance-Covariance Matrix for Slope Coefficients (Excluding Intercept)
def assemble_varBeta(pi_hat, X_with_const):
"""
Constructs the worldwide variance-covariance matrix (varBeta) for the slope coefficients
estimated from a sequence of binary logistic regressions.
Parameters:
- pi_hat : Array of form (n, Okay−1), fitted chances for every binary mannequin
- X_with_const : Array of form (n, p+1), design matrix together with intercept
Returns:
- varBeta : Array of form ((Okay−1)*p, (Okay−1)*p), block matrix of variances and covariances
throughout binary fashions (intercept excluded)
"""
# Guarantee enter is a NumPy array
X = X_with_const.values if hasattr(X_with_const, 'values') else np.asarray(X_with_const)
n, p1 = X.form # p1 = p + 1 (together with intercept)
p = p1 - 1
K_1 = pi_hat.form[1]
# Initialize world variance-covariance matrix
varBeta = np.zeros((K_1 * p, K_1 * p))
for j in vary(K_1):
pi_j = pi_hat[:, j]
Wj = np.diag(pi_j * (1 - pi_j)) # Diagonal weight matrix for mannequin j
XtWjX_inv = np.linalg.pinv(X.T @ Wj @ X)
# Take away intercept (exclude first row/column)
var_block_diag = XtWjX_inv[1:, 1:]
row_start = j * p
row_end = (j + 1) * p
varBeta[row_start:row_end, row_start:row_end] = var_block_diag
for l in vary(j + 1, K_1):
pi_l = pi_hat[:, l]
Wml = np.diag(pi_l - pi_j * pi_l)
Wl = np.diag(pi_l * (1 - pi_l))
XtWlX_inv = np.linalg.pinv(X.T @ Wl @ X)
# Compute off-diagonal block
block_cov = (XtWjX_inv @ (X.T @ Wml @ X) @ XtWlX_inv)[1:, 1:]
col_start = l * p
col_end = (l + 1) * p
# Fill symmetric blocks
varBeta[row_start:row_end, col_start:col_end] = block_cov
varBeta[col_start:col_end, row_start:row_end] = block_cov.T
return varBeta
# Compute the worldwide variance-covariance matrix
varBeta = assemble_varBeta(pi_hat, X_with_const)
# Show form and preview
print("Form of varBeta:", varBeta.form) # Anticipated: ((Okay−1) * p, (Okay−1) * p)
print("Preview of varBeta:n", varBeta[:5, :5]) # Show top-left nook
# Fill Diagonal Blocks of the World Variance-Covariance Matrix (Excluding Intercept)
def fill_varBeta_diagonal(varBeta, var_hat):
"""
Fills the diagonal blocks of the worldwide variance-covariance matrix varBeta utilizing
the person covariance matrices from every binary mannequin (excluding intercept phrases).
Parameters:
- varBeta : World variance-covariance matrix of form ((Okay−1)*p, (Okay−1)*p)
- var_hat : Listing of (p+1 × p+1) variance-covariance matrices (together with intercept)
Returns:
- varBeta : Up to date matrix with diagonal blocks crammed (intercept eliminated)
"""
K_1 = len(var_hat) # Variety of binary fashions
p = var_hat[0].form[0] - 1 # Variety of predictors (excluding intercept)
for m in vary(K_1):
block = var_hat[m][1:, 1:] # Take away intercept from variance block
row_start = m * p
row_end = (m + 1) * p
varBeta[row_start:row_end, row_start:row_end] = block
return varBeta
# Flatten the slope coefficients (excluding intercept) into betaStar
betaStar = beta_hat[:, 1:].flatten()
# Fill the diagonal blocks of the worldwide variance-covariance matrix
varBeta = fill_varBeta_diagonal(varBeta, var_hat)
# Output diagnostics
print("Form of varBeta after diagonal fill:", varBeta.form) # Anticipated: ((Okay−1)*p, (Okay−1)*p)
print("Preview of varBeta after diagonal fill:n", varBeta[:5, :5]) # Prime-left preview
Building of the (okay – 2)p x (okay – I)p distinction matrix.
import numpy as np
def construct_D(K_1, p):
"""
Constructs the distinction matrix D of form ((Okay−2)*p, (Okay−1)*p) used within the Wald check
for assessing the proportional odds assumption in ordinal logistic regression.
Parameters:
- K_1 : int — variety of binary fashions, i.e., Okay−1 thresholds
- p : int — variety of explanatory variables (excluding intercept)
Returns:
- D : numpy array of form ((Okay−2)*p, (Okay−1)*p), encoding variations between
successive β_j slope vectors (excluding intercepts)
"""
D = np.zeros(((K_1 - 1) * p, K_1 * p)) # Initialize D matrix
I = np.eye(p) # Identification matrix for block insertion
# Fill every row block with I and -I at applicable positions
for i in vary(K_1 - 1): # i = 0 to (Okay−2)
for j in vary(K_1): # j = 0 to (Okay−1)
if j == i:
temp = I # Constructive identification block
elif j == i + 1:
temp = -I # Detrimental identification block
else:
temp = np.zeros((p, p)) # Zero block in any other case
row_start = i * p
row_end = (i + 1) * p
col_start = j * p
col_end = (j + 1) * p
D[row_start:row_end, col_start:col_end] += temp
return D
# Assemble and examine D
D = construct_D(len(thresholds), len(explanatory_vars))
print("Form of D:", D.form) # Anticipated: ((Okay−2)*p, (Okay−1)*p)
print("Preview of D:n", D[:5, :5]) # Prime-left nook of D
Compute the Wald Statistic for Testing the Proportional Odds Assumption
def wald_statistic(D, betaStar, varBeta):
"""
Computes the Wald chi-square statistic X² to check the proportional odds assumption.
Parameters:
- D : (Okay−2)p × (Okay−1)p matrix that encodes linear contrasts between slope coefficients
- betaStar : Flattened vector of estimated slopes (excluding intercepts), form ((Okay−1)*p,)
- varBeta : World variance-covariance matrix of form ((Okay−1)*p, (Okay−1)*p)
Returns:
- X2 : Wald check statistic (scalar)
"""
Db = D @ betaStar # Linear contrasts of beta coefficients
V = D @ varBeta @ D.T # Variance of the contrasts
inv_V = np.linalg.inv(V) # Pseudo-inverse ensures numerical stability
X2 = float(Db.T @ inv_V @ Db) # Wald statistic
return X2
# Assumptions:
K_1 = len(binary_models) # Variety of binary fashions (Okay−1)
p = len(explanatory_vars) # Variety of explanatory variables (excluding intercept)
# Assemble distinction matrix D
D = construct_D(K_1, p)
# Compute the Wald statistic
X2 = wald_statistic(D, betaStar, varBeta)
# Levels of freedom: (Okay−2) × p
ddl = (K_1 - 1) * p
# Compute the p-value from the chi-square distribution
from scipy.stats import chi2
pval = 1 - chi2.cdf(X2, ddl)
# Output outcomes
print(f"Wald statistic X² = {X2:.4f}")
print(f"Levels of freedom = {ddl}")
print(f"p-value = {pval:.4g}")