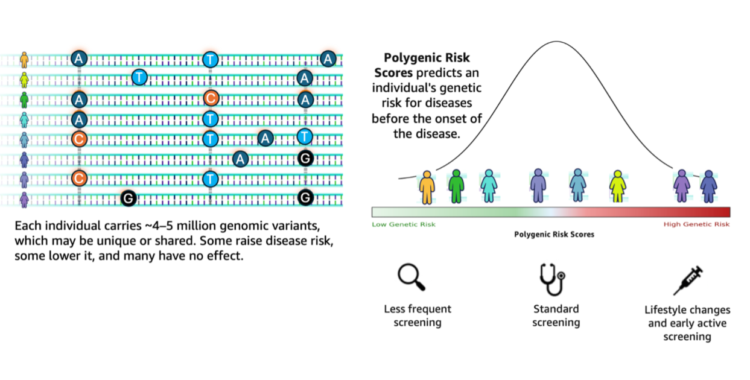
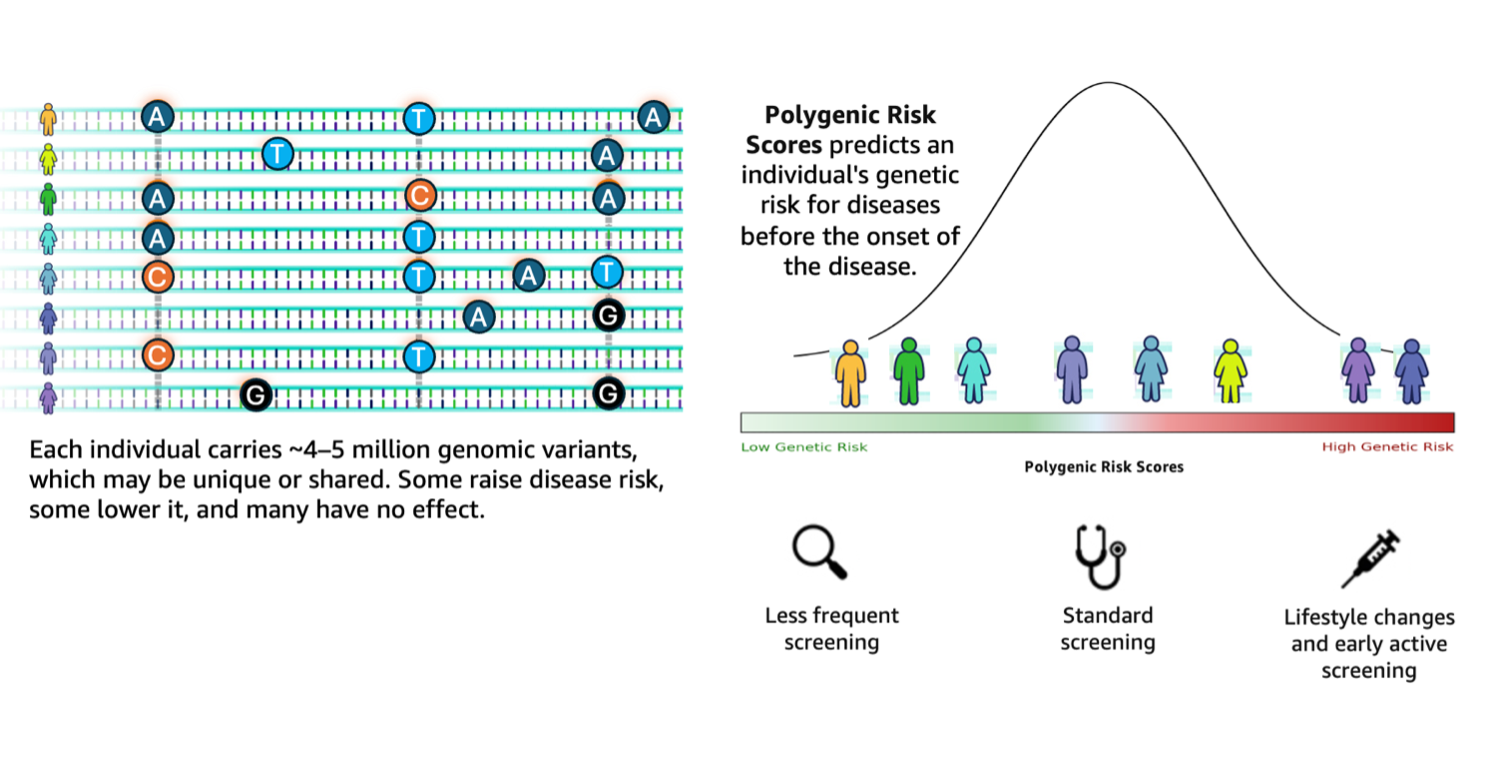
Genomic analysis stands at a transformative crossroads the place the exponential progress of sequencing information calls for equally subtle analytical capabilities. Based on the 1000 Genomes Challenge, a typical human genome differs from the reference at 4.1–5.0 million websites, with most variants being SNPs and quick indels. These variants, when aggregated throughout people, contribute to variations in illness susceptibility captured via polygenic threat scores (PRS). Genomic evaluation workflows wrestle to translate such large-scale variant information into actionable insights. They continue to be fragmented, requiring researchers to manually orchestrate complicated pipelines involving variant annotation, high quality filtering, and integration with exterior databases comparable to ClinVar.
AWS HealthOmics workflows together with Amazon S3 tables and Amazon Bedrock AgentCore collectively present a transformative resolution to those challenges. HealthOmics workflows assist the seamless integration of annotating Variant Name Format (VCF) recordsdata with insightful ontologies. Subsequently, the VEP-annotated VCF recordsdata should be remodeled into structured datasets saved in optimized S3 tables to enhance question efficiency throughout massive variant cohorts. The Strands Brokers SDK operating on Amazon Bedrock AgentCore supplies a safe and scalable AI agent utility in order that researchers can work together with complicated genomic datasets with out specialised question experience.
On this weblog submit, we present you the way agentic workflows can speed up the processing and interpretation of genomics pipelines at scale with a pure language interface. We show a complete genomic variant interpreter agent that mixes automated information processing with clever evaluation to deal with the complete workflow from uncooked VCF file ingestion to conversational question interfaces. Most significantly, this resolution removes the technical experience barrier that has historically restricted genomic evaluation to specialised bioinformaticians. This allows medical researchers to add uncooked VCF recordsdata and instantly ask questions like ‘Which sufferers have pathogenic variants in BRCA1?’ or ‘Present me drug resistance variants on this cohort’. The code for this resolution is offered within the open-source toolkit repository of starter brokers for all times sciences on AWS.
Understanding variant annotation in genomic evaluation
The inspiration of genomic variant interpretation depends on complete annotation pipelines that join uncooked genetic variants to organic and medical context. Variant Impact Predictor (VEP) and ClinVar symbolize two important parts in fashionable genomic evaluation workflows, every offering complementary info that researchers should combine to derive significant insights.
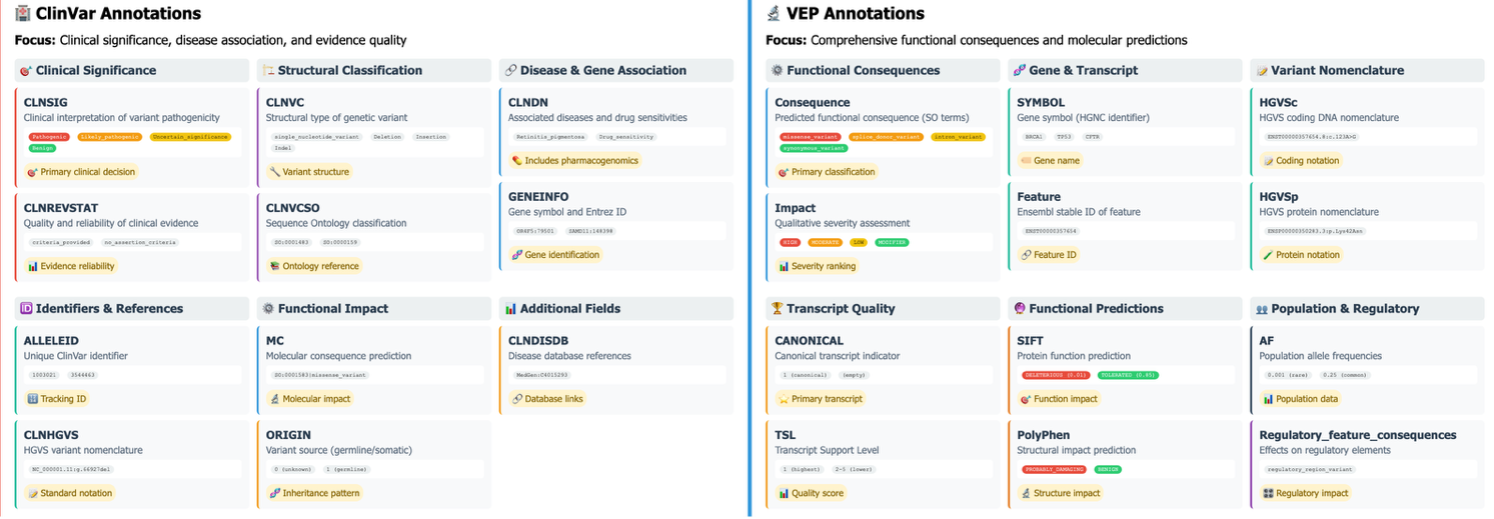
The comparative visualization illustrates the distinct but complementary annotation capabilities of ClinVar and VEP for genomic variant interpretation. ClinVar annotations (left) focus totally on medical significance evaluation, offering curated pathogenicity classifications (CLNSIG), proof high quality metrics (CLNREVSTAT), and illness associations (CLNDN) straight related to medical decision-making. VEP annotations (proper) ship complete practical info together with consequence varieties (missense_variant, synonymous_variant, intron_variant), influence severity classifications (HIGH, MODERATE, LOW, MODIFIER), gene symbols, and transcript-specific results with detailed positional info.
Present annotation workflow challenges
Variant annotation workflows sometimes observe a sequential course of that features:
- Preliminary VCF processing: Uncooked variant name format (VCF) recordsdata from sequencing programs require preprocessing to normalize illustration and filter low-quality calls.
- VEP annotation: Operating the Variant Impact Predictor instrument requires substantial computational assets, particularly for entire genome sequencing information with thousands and thousands of variants per pattern. VEP evaluation can take 2-8 hours for a single genome relying on obtainable compute assets and annotation depth.
- ClinVar integration: Medical annotations should be retrieved from ClinVar and matched to variants via a separate course of, requiring database lookups and format conversions.
- Multi-sample integration: Creating cohort-level analyses requires complicated becoming a member of operations throughout samples, sometimes carried out with specialised instruments that generate massive, flat recordsdata tough to question effectively.
- Interpretation: Scientists should then use varied instruments to filter, kind, and analyze the annotated information—a course of that always requires customized scripts and vital bioinformatics experience. This technical bottleneck implies that medical researchers can’t independently discover their genomic information, creating delays of days or even weeks between asking a organic query and receiving a solution.
Dataset complexity and scale
The size of genomic variant evaluation is exemplified by datasets just like the 1000 Genomes Section 3 Reanalysis with DRAGEN, which comprises:
- Over 2,500 particular person samples from various populations
- Roughly 85 million distinctive variants throughout all samples
- A number of annotation variations (DRAGEN 3.5, 3.7, 4.0, and 4.2) that should be reconciled
- Complicated structural variants alongside SNPs and indels
This complexity creates vital bottlenecks in conventional evaluation pipelines that depend on flat file processing and handbook integration steps.
Resolution overview
Constructing genomic cohorts or computing PRS throughout a number of sufferers calls for vital compute assets to generate joint variant name tables and complete annotations utilizing instruments just like the Variant Impact Predictor (VEP). Most critically, these workflows create a technical barrier the place solely bioinformaticians with SQL experience and deep understanding of variant file codecs can extract significant insights, leaving medical researchers depending on specialised technical groups for primary genomic queries.
The transformative benefit of our AI-powered strategy lies in democratizing genomic evaluation via pure language interplay. Whereas conventional VEP pipelines require days of technical experience to reply medical questions like ‘Which sufferers have high-impact variants in drug resistance genes?’, with our resolution researchers can ask these questions conversationally and obtain solutions in minutes. This represents a shift from technical dependency to self-service genomic insights in order that medical researchers, tumor boards, and genomics groups to straight discover their information with out ready for bioinformatics assist.
Our resolution demonstrates a generative AI-powered genomics variant interpreter agent that mixes automated information processing with clever pure language evaluation. The structure addresses the complete genomic evaluation workflow, from uncooked VCF file ingestion to conversational question interfaces.
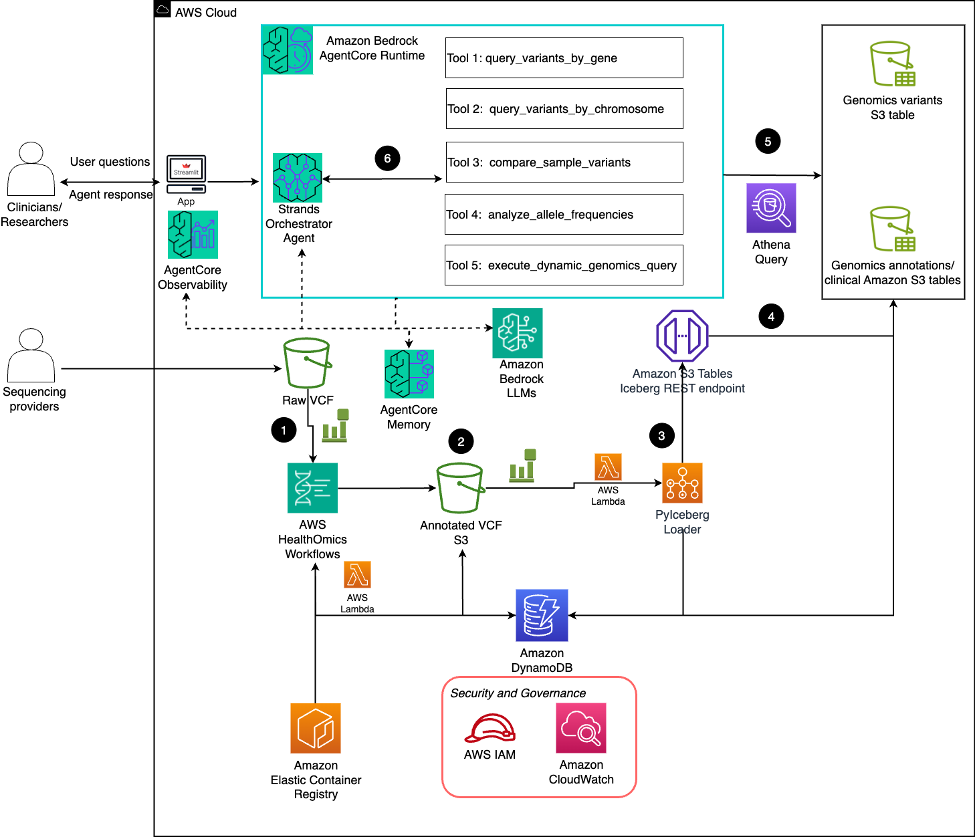
The answer follows six key steps that rework uncooked genomic information into actionable insights:
- Uncooked VCF processing: Uncooked VCF recordsdata from sequencing suppliers are uploaded to Amazon S3 storage and set off AWS Lambda features via S3 occasion notifications, which orchestrate AWS HealthOmics workflows.
- VEP annotation: AWS HealthOmics workflows mechanically course of uncooked VCF recordsdata utilizing the Variant Impact Predictor (VEP), enriching variants with practical predictions and medical annotations in parallel earlier than storing the annotated outcomes again to S3.
- Occasion coordination: Amazon EventBridge screens workflow completion and triggers Lambda features that replace job standing in Amazon DynamoDB and AWS Batch Fargate compute atmosphere transforms VEP annotated VCF recordsdata and ClinVar annotations into Iceberg format as PyIceberg module
- Knowledge group: PyIceberg loader interacts with the Amazon S3 Tables Iceberg Relaxation Endpoint. Amazon S3 Tables connects registers the desk metadata in AWS Glue Knowledge Catalog. Schema info (columns, information varieties, partitions) will get catalogued for annotated VCF and ClinVar annotations. It additionally establishes analytics connector for downstream analytics.
- SQL-powered evaluation: Amazon Athena supplies SQL-based querying capabilities over the genomic information via columnar storage format, enabling large-scale evaluation with supreme question responses throughout thousands and thousands of variants.
- Pure language interplay: The Strands orchestrator agent, powered by Amazon Bedrock LLMs on AgentCore Runtime, supplies a pure language interface via 5 specialised instruments that execute Athena queries:
- query_variants_by_gene: Retrieves variants related to particular genes
- query_variants_by_chromosome: Facilitates chromosome-specific variant evaluation
- compare_sample_variants: Allows comparative genomics throughout affected person samples
- analyze_allele_frequencies: Supplies inhabitants genetics insights
- execute_dynamic_genomics_query: Helps versatile, ad-hoc evaluation requests
The structure contains complete safety controls via AWS IAM for fine-grained entry administration and Amazon CloudWatch for monitoring. The automated, event-driven pipeline helps scalable parallel processing of VCF recordsdata that mechanically adapts to rising genomic datasets whereas sustaining constant annotation high quality and analytical capabilities.
Amazon S3 Tables with PyIceberg: Remodeling VCF to a structured cohort
Amazon S3 Tables with PyIceberg transforms VEP-annotated VCF recordsdata right into a structured cohort, queryable datasets optimized for AI-driven evaluation. This creates the info basis for pure language interfaces to effectively work together with complicated genomic information.
PyIceberg creates Apache Iceberg tables in S3 Tables format, present the next advantages:
- Optimum queries: The agent can carry out complicated genomic queries throughout thousands and thousands of variants with minimal latency via optimized columnar storage, reworking analyses that beforehand required hours of SQL growth and execution into on the spot conversational responses.
- Wealthy annotation entry: The VEP and ClinVar annotations develop into straight queryable via SQL by way of Amazon Athena, permitting the AI agent to extract particular genomic insights
- Cohort-level evaluation: The structured Iceberg format (PyIceberg) helps environment friendly comparisons throughout affected person cohorts for population-level queries via pure language.
The separation of variant information from annotation information in S3 Tables creates a great basis for AI-driven analytics as a result of genomics variants S3 tables comprise core positional info that brokers can quickly filter, and the annotations/medical S3 tables home the wealthy practical and medical context wanted for interpretation. With this construction, the Strands agent can assemble focused queries that exactly reply person questions via the AWS Glue Knowledge Catalog Connector.
This conversion from uncooked VCF recordsdata to structured tables is what makes it potential for researchers to question complicated genomic datasets conversationally via the Strands orchestrator agent [KM1] on Amazon Bedrock AgentCore.
Clever genomic evaluation with Strands Brokers and AgentCore Runtime
The conversational interface represents the core innovation of our genomics AI resolution, constructed utilizing the Strands Brokers SDK and deployed on Amazon Bedrock AgentCore Runtime. This subtle AI agent understands complicated genomic ideas and interprets pure language queries into applicable analytical operations towards the structured genomic datasets.
AgentCore Runtime is a safe, serverless runtime purpose-built for deploying and scaling dynamic AI brokers and instruments. This resolution presents a number of key benefits for genomic evaluation:
- Mannequin and framework flexibility: AgentCore providers are composable and work with open supply or customized framework and fashions, each in and out of doors of Amazon Bedrock
- Multi-hour agentic workloads: Helps long-running workloads as much as 8 hours and payloads as much as 100MB
- Safety: Devoted microVMs for every person session with full isolation
- Enterprise-grade integration: Constructed-in authentication by way of AgentCore Id with AWS IAM
- Observability: Complete tracing of agent reasoning and gear invocations
- Non-public useful resource entry: Connectivity to databases and APIs inside Amazon Digital Non-public Cloud
- Sooner time-to-market: Accelerated deployment and growth cycles for AI agent options
For detailed info on Amazon Bedrock AgentCore capabilities, consult with the Amazon Bedrock AgentCore documentation.
Strands Brokers present a sturdy basis for constructing domain-specific AI brokers with specialised capabilities via a model-driven strategy that orchestrates genomic evaluation instruments utilizing an agentic loop idea. This iterative reasoning framework allows brokers to dynamically choose and execute applicable instruments primarily based on evaluation necessities. Our genomic variant interpreter implements 5 key instruments that leverage the structured information created by Amazon S3 Tables:
- Variant querying: Interprets gene-based questions into exact Athena SQL queries that retrieve related variants.
- Chromosome evaluation: Allows region-specific genomic interrogation via pure language.
- Pattern comparability: Facilitates cross-patient genomic evaluation with out requiring SQL joins.
- Inhabitants frequency evaluation: Contextualizes findings towards reference datasets like 1000 Genomes.
- Dynamic question era: Converts complicated pure language requests into optimized SQL.
Pure language queries
The agent demonstrates outstanding functionality in dealing with various question varieties. Within the conventional mannequin medical researchers should watch for bioinformatics groups to write down customized scripts and run complicated analyses. As a substitute of spending days crafting SQL queries and wrestling with VCF file codecs, researchers can now discover their genomic information as naturally as having a dialog with a genomics professional.
Cohort-level evaluation
Consumer: "Summarize as a desk the full variety of variants and pathogenicity per affected person on this cohort?"
For this question, the agent:
- Makes use of the execute_dynamic_genomics_query instrument.
- Analyzes variant information throughout the cohort of samples.
- Generates a complete cohort abstract with affected person counts and variant statistics.
- Presents findings in a structured and tabular format abstract.
Cohort-level frequency evaluation
Consumer: "Present me the allelic frequencies of shared pathogenic or seemingly pathogenic variants on this cohort and 1000 genomes?"
The agent interprets this into queries that:
- Retrieve the checklist of pathogenic variants for the affected person by operating the execute_dynamic_genomics_query and analyze_allele_frequencies instrument.
- Filter for clinically related pathogenic variants.
- Extract illness stage info from ClinVar and allele frequencies from VEP.
- Current outcomes with related context.
Comorbidity threat affiliation
Consumer: " That are these sufferers have variant in ADRA2A gene at chr10:111079820 and, does these sufferers have any extra excessive influence variants linked with statin or insulin resistance? "
For this question, the agent:
- Searches for extra threat variants in drug resistance pathways for a particular illness context.
- Join with medical significance at particular person affected person stage for comorbidity.
- Present medical implications of joint medical and drug resistance pathways.
This pure language interface minimizes the necessity for researchers to grasp complicated SQL syntax or perceive the underlying information buildings, democratizing entry to genomic insights throughout medical and analysis groups no matter their technical background.
Superior analytic processing
Along with queries, the genomics variant interpreter agent demonstrates superior analytical capabilities that stretch past primary variant identification. Researchers can discover complicated questions that historically required days of research.
Medical resolution assist
Consumer: " Carry out an intensive evaluation on affected person NA21144 and supply me the chance stratification for this affected person"
For this question, the agent:
- Analyzes variants in illness pathways genes, pharmacogenomics, and supplies evidence-based suggestions.
- Performs threat stratification by combining variant influence predictions with medical significance classifications.
- Identifies variants of unsure significance.
- Flags high-impact variants in clinically related genes.
Pharmacogenomics guided-dosing technique
Researchers can leverage the agent for classy pharmacogenomics pathway analyses throughout massive cohorts via queries like:
Consumer: " Which main drug-related pathways are considerably enriched with genetic variants on this affected person cohort? Present me essentially the most impactful pharmacogenomic pathways and related affected person IDs "
This permits exploration of variant frequency distributions, consequence kind patterns, and gene-level variant burdens throughout totally different populations—all via conversational interfaces with out complicated SQL or bioinformatics pipelines.
Advantages and limitation
The answer helps to unravel the present challenges:
| Challenges | Options |
| Preliminary VCF processing – Low-quality calls | The agent mechanically prechecks high quality calls of variants earlier than making variant interpretation choices |
| VEP annotation at scale | The answer automates VCF annotation at scale of 20 in batches makes use of proper compute useful resource to realize the suitable efficiency. |
| ClinVar integration | The agent assess the question context and joint-query will probably be constructed dynamically primarily based on the person curiosity. |
| Multi-sample integration | Amazon S3 Tables integration in Iceberg format makes the cohort of VCF recordsdata to question with supreme efficiency. |
| Genomics interpretation | The agent understands the context and person curiosity to make the knowledgeable choices fastidiously motive out primarily based on the suitable evidences from the annotations and inhouse. |
The answer has the next limitations:
- Lambda Runtime constraints: The present implementation makes use of AWS Lambda for VCF/GVCF processing, which has a most execution time of quarter-hour. This constraint could also be inadequate for loading massive VCF recordsdata or particularly massive GVCF recordsdata into Iceberg S3 Tables, as these operations can take considerably longer than the Lambda timeout restrict. For manufacturing workloads with massive genomic datasets, think about using AWS HealthOmics workflows, AWS Batch, ECS duties, or EC2 cases with longer execution occasions to deal with the info loading course of.
- Schema optimization trade-offs: The schema implementation makes use of pattern and chromosome partitioning, which is optimized for patient-level evaluation. Nonetheless, cohort-level evaluation sometimes requires totally different partitioning methods and schema designs to realize optimum efficiency at scale. Making each patient-level and cohort-level analytics performant inside a single schema turns into more and more difficult as cohort sizes develop past a whole bunch of samples. For big-scale cohort research (1000’s to tens of 1000’s of samples), contemplate implementing separate schemas or materialized views optimized for particular analytical patterns, or discover denormalized buildings that higher assist population-level queries.
Future technological evolution
The answer’s modular structure establishes a basis for continued innovation in AI-powered genomic evaluation. Future variations may combine extra annotation databases, exterior APIs, and assist multi-modal evaluation combining genomic information with medical data and imaging. Area-specific fine-tuning on genomic information may additional enhance interpretation accuracy, whereas integration with digital well being data would offer point-of-care genomic insights.
A very promising route is multi-agent collaboration in pharmaceutical R&D, the place this genomics variant interpreter agent may work alongside specialised brokers for drug profiling, goal identification, literature proof, and speculation era. This collaborative agent framework can dramatically speed up drug discovery pipelines by connecting variant-level insights on to therapeutic growth, streamlining the interpretation from genetic findings to medical purposes.
Conclusion
This next-generation genomics agentic AI resolution represents a basic transformation in how researchers and clinicians work together with genomic information. By seamlessly integrating AWS HealthOmics for automated variant annotation and information transformation with Amazon Bedrock AgentCore for clever interpretation, we’ve created a complete resolution that addresses the complete genomic evaluation workflow.
The mix of automated VEP annotation workflows, S3 Tables for reworking VCF information into queryable Iceberg tables, and Strands Brokers on Amazon Bedrock AgentCore for pure language interplay creates a system that minimizes conventional obstacles between variant annotation, information processing, and medical interpretation. By automating complicated technical processes and offering intuitive interplay strategies, researchers can now concentrate on organic questions moderately than technical implementation particulars.
As genomic information continues to develop exponentially and medical purposes develop into more and more subtle, programs like this can develop into important infrastructure for advancing precision medication and accelerating scientific discovery. The answer demonstrated with the 1000 Genomes Section 3 Reanalysis dataset exhibits how even large-scale genomic cohorts could be analyzed via easy conversational interfaces, democratizing entry to superior genomic insights.
The code for this resolution is offered on the Life sciences brokers toolkit, and we encourage you to discover and construct upon this template. For examples to get began with Amazon Bedrock AgentCore, try the Amazon Bedrock AgentCore repository.
In regards to the authors
 Edwin Sandanaraj is a genomics options architect at AWS. With a PhD in neuro-oncology and greater than 20 years of expertise in healthcare genomics information administration and evaluation, he brings a wealth of information to speed up precision genomics efforts in Asia-Pacific and Japan. He has a passionate curiosity in medical genomics and multi-omics to speed up precision care utilizing cloud-based options.
Edwin Sandanaraj is a genomics options architect at AWS. With a PhD in neuro-oncology and greater than 20 years of expertise in healthcare genomics information administration and evaluation, he brings a wealth of information to speed up precision genomics efforts in Asia-Pacific and Japan. He has a passionate curiosity in medical genomics and multi-omics to speed up precision care utilizing cloud-based options.
 Hasan Poonawala is a Senior AI/ML Options Architect at AWS, working with Healthcare and Life Sciences prospects. Hasan helps design, deploy and scale Generative AI and Machine studying purposes on AWS. He has over 15 years of mixed work expertise in machine studying, software program growth and information science on the cloud. In his spare time, Hasan likes to discover nature and spend time with family and friends.
Hasan Poonawala is a Senior AI/ML Options Architect at AWS, working with Healthcare and Life Sciences prospects. Hasan helps design, deploy and scale Generative AI and Machine studying purposes on AWS. He has over 15 years of mixed work expertise in machine studying, software program growth and information science on the cloud. In his spare time, Hasan likes to discover nature and spend time with family and friends.
 Charlie Lee is genomics business lead for Asia-Pacific and Japan at AWS and has a PhD in laptop science with a concentrate on bioinformatics. An business chief with greater than 20 years of expertise in bioinformatics, genomics, and molecular diagnostics, he’s captivated with accelerating analysis and enhancing healthcare via genomics with cutting-edge sequencing applied sciences and cloud computing.
Charlie Lee is genomics business lead for Asia-Pacific and Japan at AWS and has a PhD in laptop science with a concentrate on bioinformatics. An business chief with greater than 20 years of expertise in bioinformatics, genomics, and molecular diagnostics, he’s captivated with accelerating analysis and enhancing healthcare via genomics with cutting-edge sequencing applied sciences and cloud computing.