This publish is co-authored with the Biomni group from Stanford.
Biomedical researchers spend roughly 90% of their time manually processing large volumes of scattered info. That is evidenced by Genentech’s problem of processing 38 million biomedical publications in PubMed, public repositories just like the Human Protein Atlas, and their inner repository of a whole bunch of thousands and thousands of cells throughout a whole bunch of illnesses. There’s a fast proliferation of specialised databases and analytical instruments throughout totally different modalities together with genomics, proteomics, and pathology. Researchers should keep present with the big panorama of instruments, leaving much less time for the hypothesis-driven work that drives breakthrough discoveries.
AI brokers powered by basis fashions provide a promising resolution by autonomously planning, executing, and adapting advanced analysis duties. Stanford researchers constructed Biomni that exemplifies this potential. Biomni is a general-purpose biomedical AI agent that integrates 150 specialised instruments, 105 software program packages, and 59 databases to execute refined analyses equivalent to gene prioritization, drug repurposing, and uncommon illness prognosis.
Nonetheless, deploying such brokers in manufacturing requires strong infrastructure able to dealing with computationally intensive workflows and a number of concurrent customers whereas sustaining safety and efficiency requirements. Amazon Bedrock AgentCore is a set of complete companies to deploy and function extremely succesful brokers utilizing any framework or mannequin, with enterprise-grade safety and scalability.
On this publish, we present you find out how to implement a analysis agent utilizing AgentCore with entry to over 30 specialised biomedical database instruments from Biomni, thereby accelerating scientific discovery whereas sustaining enterprise-grade safety and manufacturing scale. The code for this resolution is offered within the open-source toolkit repository of starter brokers for all times sciences on Amazon Internet Companies (AWS). The step-by-step instruction helps you deploy your individual instruments and infrastructure, together with AgentCore elements, and examples.
Prototype-to-production complexity hole
Shifting from an area biomedical analysis prototype to a manufacturing system accessible by a number of analysis groups requires addressing advanced infrastructure challenges.
Agent deployment with enterprise safety
Enterprise safety challenges embody OAuth-based authentication, safe device sharing via scalable gateways, complete observability for analysis audit trails, and automated scaling to deal with concurrent analysis workloads. Many promising prototypes fail to succeed in manufacturing due to the complexity of implementing these enterprise-grade necessities whereas sustaining the specialised area experience wanted for correct biomedical evaluation.
Session-aware analysis context administration
Biomedical analysis workflows typically span a number of conversations and require persistent reminiscence of earlier analyses, experimental parameters, and analysis preferences throughout prolonged analysis periods. Analysis brokers should preserve contextual consciousness of ongoing tasks, keep in mind particular protein targets, experimental circumstances, and analytical preferences. All that have to be achieved whereas facilitating correct session isolation between totally different researchers and analysis tasks in a multi-tenant manufacturing atmosphere.
Scalable device gateway
Implementing a reusable device gateway that may deal with concurrent requests from analysis agent, correct authentication, and constant efficiency turns into crucial at scale. The gateway should allow brokers to find and use instruments via safe endpoints, assist brokers discover the best instruments via contextual search capabilities, and handle each inbound authentication (verifying agent identification) and outbound authentication (connecting to exterior biomedical databases) in a unified service. With out this structure, analysis groups face authentication complexity and reliability points that forestall efficient scaling.
Resolution overview
We use Strands Brokers, an open supply agent framework, to construct a analysis agent with native device implementation for PubMed biomedical literature search. We prolonged the agent’s capabilities by integrating Biomni database instruments, offering entry to over 30 specialised biomedical databases.
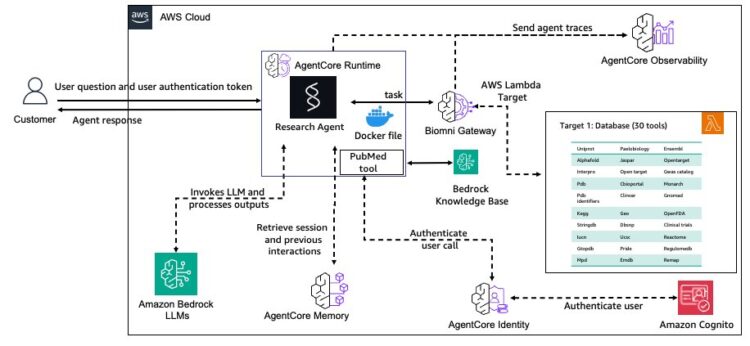
The general structure is proven within the following diagram.
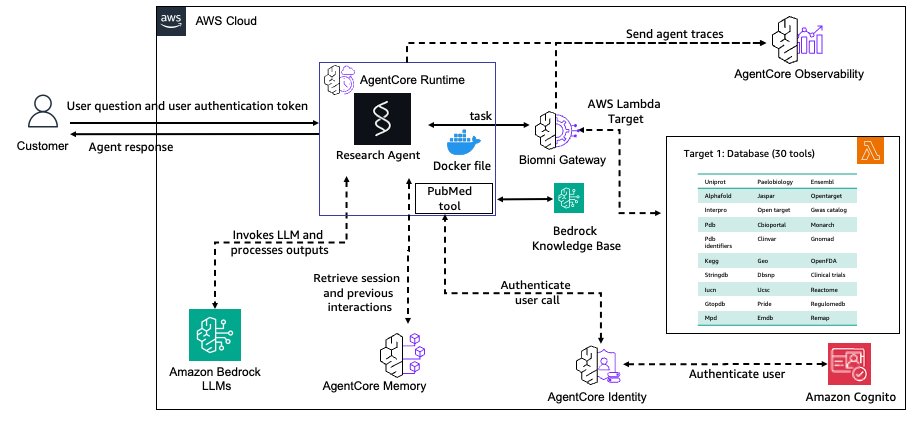
The AgentCore Gateway service centralizes Biomni database instruments as safer, reusable endpoints with semantic search capabilities. AgentCore Reminiscence service maintains contextual consciousness throughout analysis periods utilizing specialised methods for analysis context. Safety is dealt with by AgentCore Identification service, which manages authentication for each customers and power entry management. Deployment is streamlined with the AgentCore Runtime service, offering scalable, managed deployment with session isolation. Lastly, the AgentCore Observability service allows complete monitoring and auditing of analysis workflows which are crucial for scientific reproducibility.
Step 1 – Creating instruments such because the Biomni database instruments utilizing AgentCore Gateway
In real-world use circumstances, we have to join brokers to totally different information sources. Every agent would possibly duplicate the identical instruments, resulting in in depth code, inconsistent habits, and upkeep nightmares. AgentCore Gateway service streamlines this course of by centralizing instruments into reusable, safe endpoints that brokers can entry. Mixed with the AgentCore Identification service for authentication, AgentCore Gateway creates an enterprise-grade device sharing infrastructure. To present extra context to the agent with reusable instruments, we offered entry to over 30 specialised public database APIs via the Biomni instruments registered on the gateway. The gateway exposes Biomni’s database instruments via the Mannequin Context Protocol (MCP), permitting the analysis agent to find and invoke these instruments alongside native instruments like PubMed. It handles authentication, charge limiting, and error dealing with, offering a seamless analysis expertise.
We use an AWS Lambda perform to host the Biomni integration code. The Lambda perform is robotically configured as an MCP goal within the AgentCore Gateway. The Lambda perform exposes its out there instruments via the API specification (api_spec.json).
# Gateway Goal Configuration
lambda_target_config = {
"mcp": {
"lambda": {
"lambdaArn": get_ssm_parameter("/app/researchapp/agentcore/lambda_arn"),
"toolSchema": {"inlinePayload": api_spec},
}
}
}
# Create the goal
create_target_response = gateway_client.create_gateway_target(
gatewayIdentifier=gateway_id,
title="LambdaUsingSDK",
description="Lambda Goal utilizing SDK",
targetConfiguration=lambda_target_config,
credentialProviderConfigurations=[{
"credentialProviderType": "GATEWAY_IAM_ROLE"
}],
)The complete record of Biomni database instruments included on the gateway are listed within the following desk:
| Group | Software | Description |
| Protein and construction databases | UniProt | Question the UniProt REST API for complete protein sequence and purposeful info |
| AlphaFold | Question the AlphaFold Database API for AI-predicted protein construction predictions | |
| InterPro | Question the InterPro REST API for protein domains, households, and purposeful websites | |
| PDB (Protein Knowledge Financial institution) | Question the RCSB PDB database for experimentally decided protein constructions | |
| STRING | Question the STRING protein interplay database for protein-protein interplay networks | |
| EMDB (Electron Microscopy Knowledge Financial institution) | Question for 3D macromolecular constructions decided by electron microscopy | |
| Genomics and variants | ClinVar | Question NCBI's ClinVar database for clinically related genetic variants and their interpretations |
| dbSNP | Question the NCBI dbSNP database for single nucleotide polymorphisms and genetic variations | |
| gnomAD | Question gnomAD for population-scale genetic variant frequencies and annotations | |
| Ensembl | Question the Ensembl REST API for genome annotations, gene info, and comparative genomics | |
| UCSC Genome Browser | Question the UCSC Genome Browser API for genomic information and annotations | |
| Expression and omics | GEO (Gene Expression Omnibus) | Question NCBI's GEO for RNA-seq, microarray, and different gene expression datasets |
| PRIDE | Question the PRIDE database for proteomics identifications and mass spectrometry information | |
| Reactome | Question the Reactome database for organic pathways and molecular interactions | |
| Medical and drug information | cBioPortal | Question the cBioPortal REST API for most cancers genomics information and scientific info |
| ClinicalTrials.gov | Question ClinicalTrials.gov API for details about scientific research and trials | |
| OpenFDA | Question the OpenFDA API for FDA drug, system, and meals security information | |
| GtoPdb (Information to PHARMACOLOGY) | Question the Information to PHARMACOLOGY database for drug targets and pharmacological information | |
| Illness and phenotype | OpenTargets | Question the OpenTargets Platform API for disease-target associations and drug discovery information |
| Monarch Initiative | Question the Monarch Initiative API for phenotype and illness info throughout species | |
| GWAS Catalog | Question the GWAS Catalog API for genome-wide affiliation research outcomes | |
| RegulomeDB | Question the RegulomeDB database for regulatory variant annotations and purposeful predictions | |
| Specialised databases | JASPAR | Question the JASPAR REST API for transcription issue binding web site profiles and motifs |
| WoRMS (World Register of Marine Species) | Question the WoRMS REST API for marine species taxonomic info | |
| Paleobiology Database (PBDB) | Question the PBDB API for fossil incidence and taxonomic information | |
| MPD (Mouse Phenome Database) | Question the Mouse Phenome Database for mouse pressure phenotype information | |
| Synapse | Question Synapse REST API for biomedical datasets and collaborative analysis information |
The next are examples of how particular person instruments get triggered via the MCP from our check suite:
Because the device assortment grows, the agent can use built-in semantic search capabilities to find and choose instruments primarily based on the duty context. This improves agent efficiency and decreasing improvement complexity at scale. For instance, the person asks, “inform me about HER2 variant rs1136201.” As an alternative of itemizing all 30 or extra instruments from the gateway again to the agent, semantic search returns ‘n’ most related instruments. For instance, Ensembl, Gwas catalog, ClinVar, and Dbsnp to the agent. The agent now makes use of a smaller subset of instruments as enter to the mannequin to return a extra environment friendly and sooner response.
The next graphic illustrates utilizing AgentCore Gateway for device search.
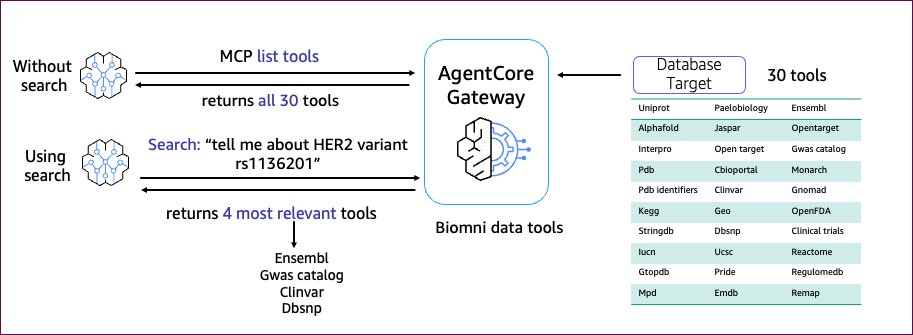
Now you can check your deployed AgentCore gateway utilizing the next check scripts and examine how semantic search narrows down the record of related instruments primarily based on the search question.
uv run assessments/test_gateway.py --prompt "What instruments can be found?"
uv run assessments/test_gateway.py --prompt "Discover details about human insulin protein" --use-searchStep 2- Strands analysis agent with an area device
The next code snippet reveals mannequin initialization, implementing the PubMed native device that’s declared utilizing the Strands @device decorator. We’ve applied the PubMed device in research_tools.py that calls PubMed APIs to allow biomedical literature search capabilities throughout the agent's execution context.
class ResearchAgent:
def __init__(
self,
bearer_token: str,
memory_hook: MemoryHook = None,
session_manager: AgentCoreMemorySessionManager = None,
bedrock_model_id: str = "us.anthropic.claude-sonnet-4-20250514-v1.0",
#bedrock_model_id: str = "openai.gpt-oss-120b-1.0", # Different
system_prompt: str = None,
instruments: Listing[callable] = None,
):
self.model_id = bedrock_model_id
# For Anthropic Sonnet 4 interleaved considering
self.mannequin = BedrockModel(
model_id=self.model_id,
additional_request_fields={
"anthropic_beta": ["interleaved-thinking-2025-05-14"],
"considering": {"sort": "enabled", "budget_tokens": 8000},
},
)
self.system_prompt = (
system_prompt
if system_prompt
else """
You're a **Complete Biomedical Analysis Agent** specialised in conducting
systematic literature opinions and multi-database analyses to reply advanced biomedical analysis
questions. Your main mission is to synthesize proof from each revealed literature
(PubMed) and real-time database queries to supply complete, evidence-based insights for
pharmaceutical analysis, drug discovery, and scientific decision-making.
Your core capabilities embody literature evaluation and extracting information from 30+ specialised
biomedical databases** via the Bioimm gateway, enabling complete information evaluation. The
database device classes embody genomics and genetics, protein construction and performance, pathways
and system biology, scientific and pharmacological information, expression and omics information and different
specialised databases.
"""
)- As well as, we applied citations that use a structured system immediate to implement numbered in-text citations [1], [2], [3] with standardized reference codecs for each tutorial literature and database queries, marking certain each information supply is correctly attributed. This permits researchers to shortly entry and reference the scientific literature that helps their biomedical analysis queries and findings.
"""
- ALWAYS use numbered in-text citations [1], [2], [3], and so on. when referencing any information supply
- Present a numbered "References" part on the finish with full supply particulars
- For educational literature: format as "1. Creator et al. Title. Journal. 12 months. ID: [PMID/DOI], out there at: [URL]"
- For database sources: format as "1. Database Identify (Software: tool_name), Question: [query_description], Retrieved: [current_date]"
- Use numbered in-text citations all through your response to assist all claims and information factors
- Every device question and every literature supply have to be cited with its personal distinctive reference quantity
- When instruments return tutorial papers, cite them utilizing the tutorial format with full bibliographic particulars
- Construction: Format every reference on a separate line with correct numbering - NO bullet factors
- Current the References part as a clear numbered record, not a complicated paragraph
- Preserve sequential numbering throughout all reference varieties in a single "References" part
"""
Now you can check your agent regionally:
uv run assessments/test_agent_locally.py --prompt "Discover details about human insulin protein"
uv run assessments/test_agent_locally.py --prompt "Discover details about human insulin protein" --use-searchStep 3 - Add Persistent Reminiscence for contextual analysis help
The analysis agent implements the AgentCore Reminiscence service with three methods: semantic for factual analysis context, user_preference for analysis methodologies, and abstract for session continuity. The AgentCore Reminiscence session supervisor is built-in with Strands session administration and retrieves related context earlier than queries and save interactions after responses. This allows the agent to recollect analysis preferences, ongoing tasks, and area experience throughout periods with out guide context re-establishment.
# Take a look at reminiscence performance with analysis conversations
python assessments/test_memory.py load-conversation
python assessments/test_memory.py load-prompt "My most popular response format is detailed explanations"Step 4 - Deploy with AgentCore Runtime
To deploy our agent, we use AgentCore Runtime to configure and launch the analysis agent as a managed service. The deployment course of configures the runtime with the agent's most important entrypoint (agent/most important.py), assigns an IAM execution position for AWS service entry, and helps each OAuth and IAM authentication modes. After deployment, the runtime turns into a scalable, serverless agent that may be invoked utilizing API calls. The agent robotically handles session administration, reminiscence persistence, and power orchestration whereas offering safe entry to the Biomni gateway and native analysis instruments.
agentcore configure --entrypoint agent/most important.py -er arn:aws:iam::<Account-Id>:position/<Function> --name researchapp<AgentName>For extra details about deploying with AgentCore Runtime, see Get began with AgentCore Runtime within the Amazon Bedrock AgentCore Developer Information.
Brokers in motion
The next are three consultant analysis situations that showcase the agent's capabilities throughout totally different domains: drug mechanism evaluation, genetic variant investigation, and pathway exploration. For every question, the agent autonomously determines which mixture of instruments to make use of, formulates acceptable sub-queries, analyzes the returned information, and synthesizes a complete analysis report with correct citations. The accompanying demo video reveals the entire agent workflow, together with instruments choice, reasoning, and response technology.
- Conduct a complete evaluation of trastuzumab (Herceptin) mechanism of motion and resistance mechanisms you’ll want:
- HER2 protein construction and binding websites
- Downstream signaling pathways affected
- Identified resistance mechanisms from scientific information
- Present scientific trials investigating mixture therapies
- Biomarkers for remedy response predictionQuery related databases to supply a complete analysis report.
- Analyze the scientific significance of BRCA1 variants in breast most cancers danger and remedy response. Examine:
- Inhabitants frequencies of pathogenic BRCA1 variants
- Medical significance and pathogenicity classifications
- Related most cancers dangers and penetrance estimates
- Therapy implications (PARP inhibitors, platinum brokers)
- Present scientific trials for BRCA1-positive sufferers
Use a number of databases to supply complete proof
The next video is an illustration of a biomedical analysis agent:
Scalability and observability
One of the vital crucial challenges in deploying refined AI brokers is ensuring they scale reliably whereas sustaining complete visibility into their operations. Biomedical analysis workflows are inherently unpredictable—a single genomic evaluation would possibly course of 1000's of recordsdata, whereas a literature evaluate might span thousands and thousands of publications. Conventional infrastructure struggles with these dynamic workloads, notably when dealing with delicate analysis information that requires strict isolation between totally different analysis tasks.On this deployment, we use Amazon Bedrock AgentCore Observability to visualise every step within the agent workflow. You should use this service to examine an agent's execution path, audit intermediate outputs, and debug efficiency bottlenecks and failures. For biomedical analysis, this stage of transparency is not only useful—it is important for regulatory compliance and scientific reproducibility.
Periods, traces, and spans type a three-tiered hierarchical relationship within the observability framework. A session accommodates a number of traces, with every hint representing a discrete interplay throughout the broader context of the session. Every hint accommodates a number of spans that seize fine-grained operations. The next screenshot sneakers the utilization of 1 agent: Variety of periods, token utilization, and error charge in manufacturing
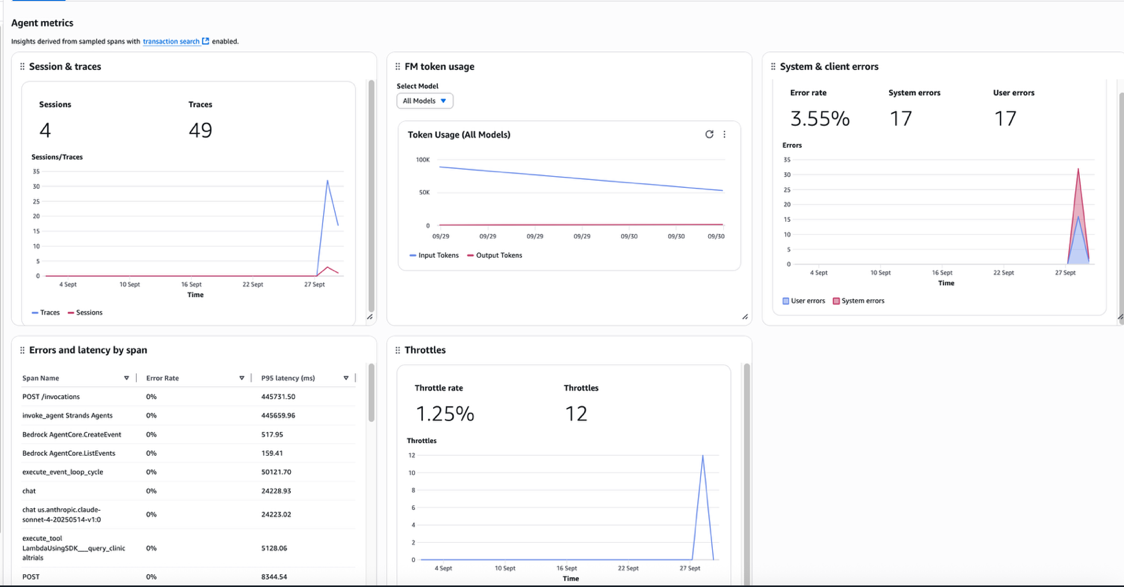
The next screenshot reveals the brokers in manufacturing and their utilization (variety of Periods, variety of invocations)
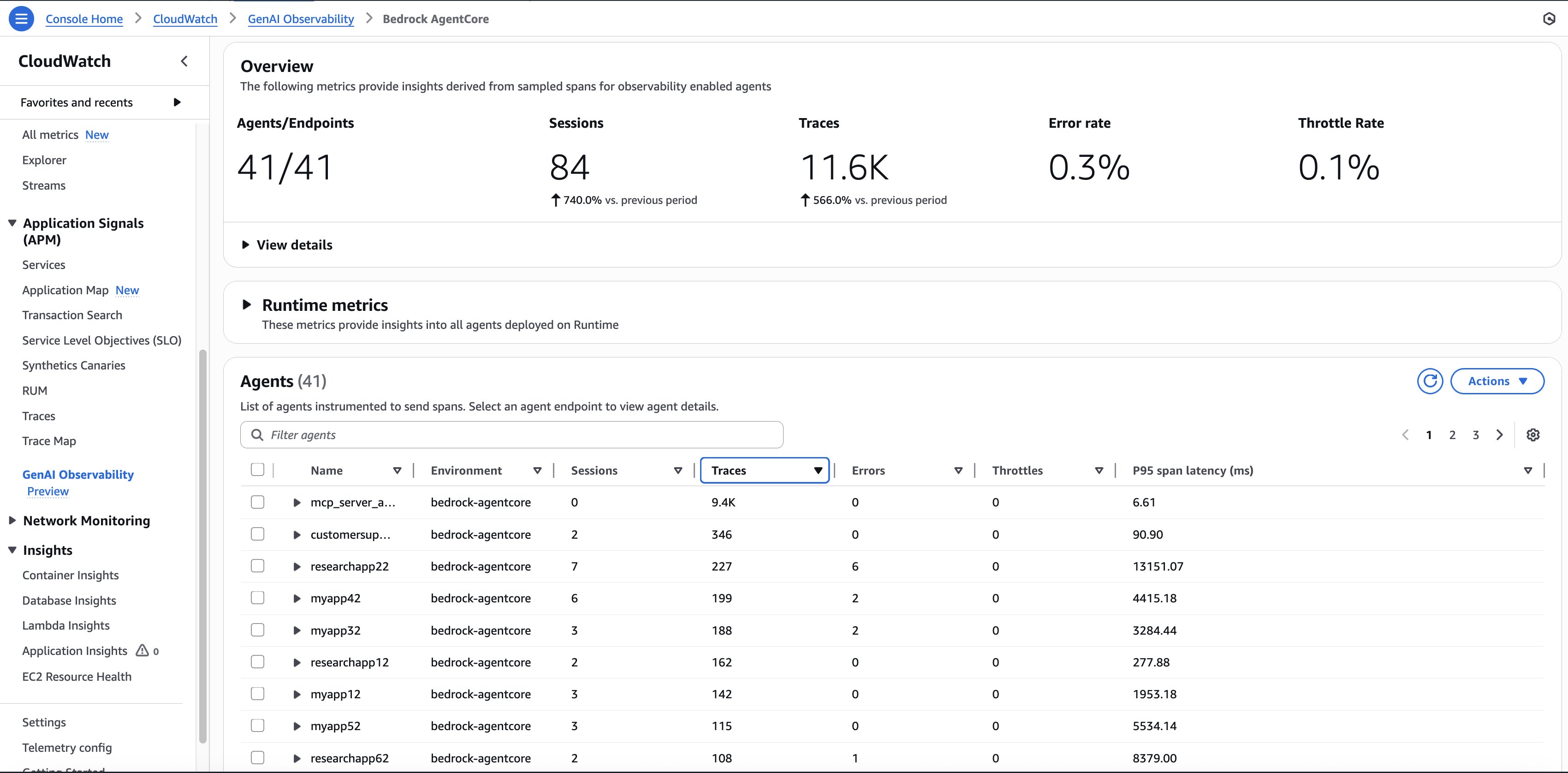
The built-in dashboards present efficiency bottlenecks and determine why sure interactions would possibly fail, enabling steady enchancment and decreasing the imply time to detect (MTTD) and imply time to restore (MTTR). For biomedical functions the place failed analyses can delay crucial analysis timelines, this fast concern decision functionality makes certain that analysis momentum is maintained.
Future route
Whereas this implementation focuses on solely a subset of instruments, the AgentCore Gateway structure is designed for extensibility. Analysis groups can seamlessly add new instruments with out requiring code adjustments through the use of the MCP protocol. Newly registered instruments are robotically discoverable by brokers permitting your analysis infrastructure to evolve alongside the quickly altering device units.
For computational evaluation that requires code execution, the AgentCore Code Interpreter service will be built-in into the analysis workflow. With AgentCore Code Interpreter the analysis agent can retrieve information and execute Python-based evaluation utilizing domain-specific libraries like BioPython, scikit-learn, or customized genomics packages.
Future extensions might assist a number of analysis brokers to collaborate on advanced tasks, with specialised brokers for literature evaluate, experimental design, information evaluation, and end result interpretation working collectively via multi-agent collaboration. Organizations can even develop specialised analysis brokers tailor-made to particular therapeutic areas, illness domains, or analysis methodologies that share the identical enterprise infrastructure and power gateway.
Wanting forward with Biomni
“Biomni right this moment is already helpful for educational analysis and open exploration. However to allow actual discovery—like advancing drug improvement—we have to transfer past prototypes and make the system enterprise-ready. Embedding Biomni into the workflows of biotech and pharma is important to show analysis potential into tangible influence.
That’s why we're excited to combine the open-source atmosphere with Amazon Bedrock AgentCore, bridging the hole from analysis to manufacturing. Wanting forward, we’re additionally enthusiastic about extending these capabilities with the Biomni A1 agent structure and the Biomni-R0 mannequin, which can unlock much more refined biomedical reasoning and evaluation. On the similar time, Biomni will stay a thriving open-source atmosphere, the place researchers and business groups alike can contribute instruments, share workflows, and push the frontier of biomedical AI along with AgentCore.”
Conclusion
This implementation demonstrates how organizations can use Amazon Bedrock AgentCore to rework biomedical analysis prototypes into production-ready methods. By integrating Biomni's complete assortment of over 150 specialised instruments via the AgentCore Gateway service, we illustrate how groups can create enterprise-grade device sharing infrastructure that scales throughout a number of analysis domains.The mixture of Biomni's biomedical instruments with the enterprise infrastructure of Bedrock AgentCore organizations can construct analysis brokers that preserve scientific rigor whereas assembly manufacturing necessities for safety, scalability, and observability. Biomni's various device assortment—spanning genomics, proteomics, and scientific databases—exemplifies how specialised analysis capabilities will be centralized and shared throughout analysis groups via a safe gateway structure.
To start constructing your individual biomedical analysis agent with Biomni instruments, discover the implementation by visiting our GitHub repository for the entire code and documentation. You'll be able to comply with the step-by-step implementation information to arrange your analysis agent with native instruments, gateway integration, and Bedorck AgentCore deployment. As your wants evolve, you may lengthen the system along with your group's proprietary databases and analytical instruments. We encourage you to hitch the rising atmosphere of life sciences AI brokers and instruments by sharing your extensions and enhancements.
In regards to the authors
 Hasan Poonawala is a Senior AI/ML Options Architect at AWS, working with Healthcare and Life Sciences prospects. Hasan helps design, deploy and scale Generative AI and Machine studying functions on AWS. He has over 15 years of mixed work expertise in machine studying, software program improvement and information science on the cloud. In his spare time, Hasan likes to discover nature and spend time with family and friends.
Hasan Poonawala is a Senior AI/ML Options Architect at AWS, working with Healthcare and Life Sciences prospects. Hasan helps design, deploy and scale Generative AI and Machine studying functions on AWS. He has over 15 years of mixed work expertise in machine studying, software program improvement and information science on the cloud. In his spare time, Hasan likes to discover nature and spend time with family and friends.
 Pierre de Malliard is a Senior AI/ML Options Architect at Amazon Internet Companies and helps prospects within the Healthcare and Life Sciences Business. He's at present primarily based in New York Metropolis.
Pierre de Malliard is a Senior AI/ML Options Architect at Amazon Internet Companies and helps prospects within the Healthcare and Life Sciences Business. He's at present primarily based in New York Metropolis.
 Necibe Ahat is a Senior AI/ML Specialist Options Architect at AWS, working with Healthcare and Life Sciences prospects. Necibe helps prospects to advance their generative AI and machine studying journey. She has a background in laptop science with 15 years of business expertise serving to prospects ideate, design, construct and deploy options at scale. She is a passionate inclusion and variety advocate.
Necibe Ahat is a Senior AI/ML Specialist Options Architect at AWS, working with Healthcare and Life Sciences prospects. Necibe helps prospects to advance their generative AI and machine studying journey. She has a background in laptop science with 15 years of business expertise serving to prospects ideate, design, construct and deploy options at scale. She is a passionate inclusion and variety advocate.
 Kexin Huang is a final-year PhD scholar in Laptop Science at Stanford College, suggested by Prof. Jure Leskovec. His analysis applies AI to allow interpretable and deployable biomedical discoveries, addressing core challenges in multi-modal modeling, uncertainty, and reasoning. His work has appeared in Nature Medication, Nature Biotechnology, Nature Chemical Biology, Nature Biomedical Engineering and high ML venues (NeurIPS, ICML, ICLR), incomes six greatest paper awards. His analysis has been highlighted by Forbes, WIRED, and MIT Know-how Assessment, and he has contributed to AI analysis at Genentech, GSK, Pfizer, IQVIA, Flatiron Well being, Dana-Farber, and Rockefeller College.
Kexin Huang is a final-year PhD scholar in Laptop Science at Stanford College, suggested by Prof. Jure Leskovec. His analysis applies AI to allow interpretable and deployable biomedical discoveries, addressing core challenges in multi-modal modeling, uncertainty, and reasoning. His work has appeared in Nature Medication, Nature Biotechnology, Nature Chemical Biology, Nature Biomedical Engineering and high ML venues (NeurIPS, ICML, ICLR), incomes six greatest paper awards. His analysis has been highlighted by Forbes, WIRED, and MIT Know-how Assessment, and he has contributed to AI analysis at Genentech, GSK, Pfizer, IQVIA, Flatiron Well being, Dana-Farber, and Rockefeller College.